Persistently Infected Covid-19 Patients: A Potential Source For New Variants
(Posted on Tuesday, February 16, 2021)
Random variation is an essential component of all living things. It drives diversity, and it is why there are so many different species. Viruses are no exception. Most viruses are experts at changing genomes to adapt to their environment. We now have evidence that the virus that causes Covid, SARS-CoV-2, not only changes, but changes in ways that are significant. This is the twelfth part of a series of articles on how the virus changes and what that means for humanity. Read the rest: part one, part two, part three, part four, part five, part six, part seven, part eight, part nine, part ten, and part eleven.
Colorized scanning electron micrograph of an apoptotic cell (red) infected with SARS-COV-2 virus particles (yellow), isolated from a patient sample. Image captured at the NIAID Integrated Research Facility (IRF) in Fort Detrick, Maryland. Credit:
BSIP/UNIVERSAL IMAGES GROUP VIA GETTY IMAGES
Previously I described the evolution of a viral variant in an immunosuppressed patient in London who was persistently infected with SARS-CoV-2. I also went into detail about the specific changes in the genomes of viruses isolated from the London patient that were identical to those identified in the variants of concern in South Africa and Brazil. Some of the same mutations are found in laboratory experiments that purposefully generate and select for virus strains resistant to antibodies in the blood of patients who have recovered from Covid-19.
These observations raise two interesting possibilities. Can it be that persistently infected, immunosuppressed Covid-19 patients are the actual source of variants? Or are the pathways to increased transmission, virulence, and immune evasion in populations independent but parallel to those occurring in immunosuppressed patients and laboratory experiments? Either way, taking a look at the viral variants that arise in infected, immunosuppressed patients can provide us a valuable window into what the future may hold. Here I describe observations of another immunosuppressed, persistently infected patient in Boston.
In November 2020, the New England Journal of Medicine published a case study of a 45-year-old man who at some point last year checked into the hospital with a bad fever and some other minor Covid-19 symptoms—henceforth known as the Boston patient. At the time, the Boston patient was undergoing treatment for severe antiphospholipid syndrome, an autoimmune disorder that causes the body to turn against its own cells. He ended up staying in the hospital for a total of 154 days, from the day he first tested positive for Covid-19 up until the day he passed away (Figure 1).
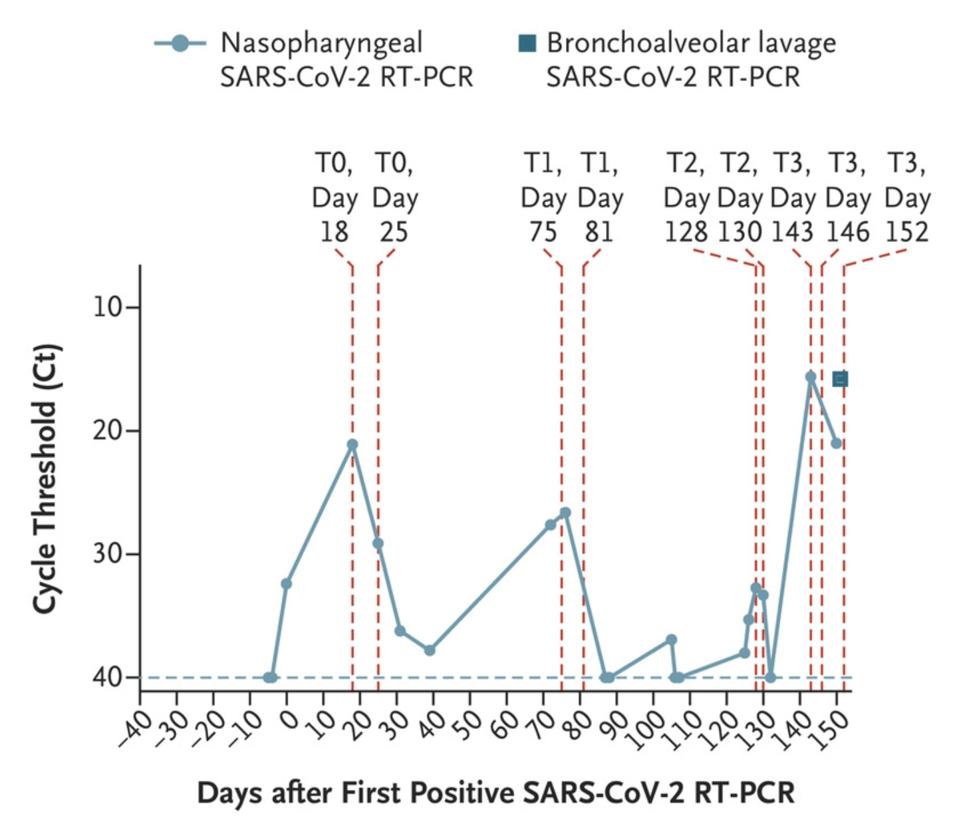
Figure 1. Nasopharyngeal and bronchoalveolar-lavage SARS-CoV-2 RT-PCR cycle threshold (Ct) values; the horizontal dashed line represents the cutoff for positivity at 40, and vertical red dashed lines represent days of viral sequencing (days 18, 25,
“PERSISTENCE AND EVOLUTION OF SARS-COV-2 IN AN IMMUNOCOMPROMISED HOST” HTTPS://WWW.NEJM.ORG/DOI/FULL/10.1056/NEJMC2031364The doctors treating the Boston patient gave him several experimental therapies over the course of his hospitalization, from the antiviral remdesivir to the Regeneron antibody cocktail, which contains two SARS-CoV-2 monoclonal antibodies. As was the case with the London patient, rather than causing the virus to subsist, these treatments evidently put more pressure on it to evolve. Also like the London patient, the man’s immune system was already weakened by a preexisting health condition, creating an ideal environment for the virus to continuously experiment with newer, better versions of itself—the result being the development of multiple mutations, as many as 24, along the spike protein (Figure 2, Figure 3).
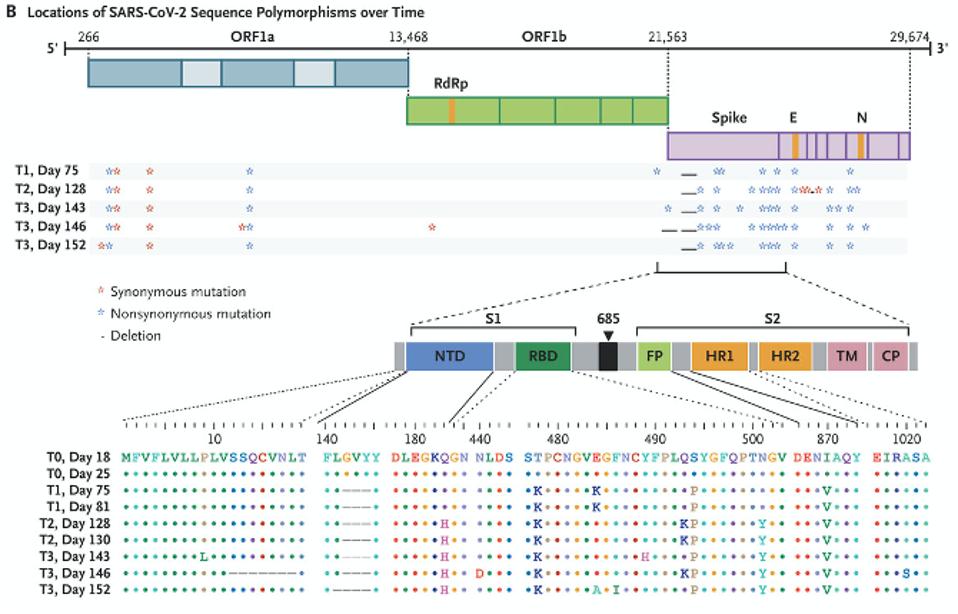
Figure 2. The mutations in the Boston patient include deletions from 12-18 in the signal peptide and N-terminal domain; deletions 142-144 in the N-terminal domain; the Q183H substitution in the N-terminal domain; N440D, T478K, E484K/A, F486I, Y489H,
“PERSISTENCE AND EVOLUTION OF SARS-COV-2 IN AN IMMUNOCOMPROMISED HOST” HTTPS://WWW.NEJM.ORG/DOI/FULL/10.1056/NEJMC2031364You may recall that the spike is what SARS-CoV-2 uses to latch onto our cells’ ACE2 receptors. In the Boston patient, almost all of the mutations that appear in this part of the virus were located in two main subregions, the N-terminal domain and the receptor binding domain. In the N-terminal domain, deletions occurred from sites 142 through 144 (Figure 4). While not much is known about the functionality or broader significance of these particular amino acids, it does appear that SARS-CoV-2 can sustain multiple deletions in the N-terminal domain and still maintain full infectivity. The London patient also had a mutation in this area—the H69-70 deletion that later appeared in the B.1.1.7, or UK, variant.
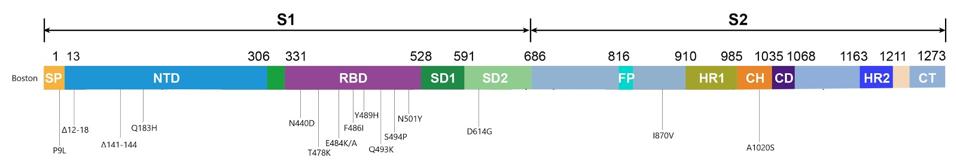
Figure 3. Visual representation of the spike protein mutations.
AUTHOR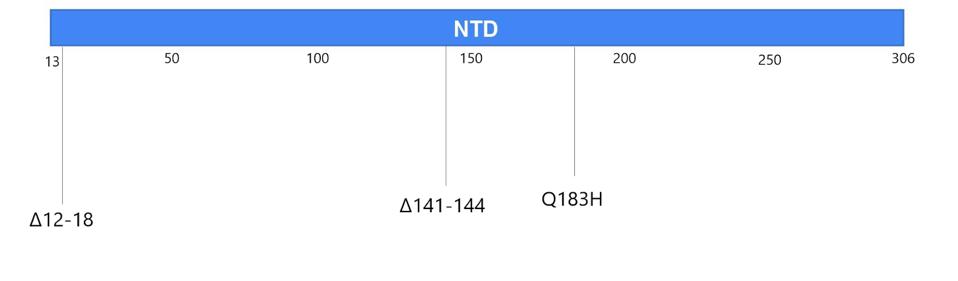
Figure 4. Visual representation of mutations to the N-terminal domain.
AUTHORThe deletions in the N-terminal domain are most likely to delete neutralizing antibody epitopes, while maintaining the function of the entire S1 protein. On the other hand, all the mutations in the receptor binding domain are point mutations, or substitutions, which may affect immunogenicity and affinity but leave existing functions intact. This region seems to be intolerant of deletions. D614G, a mutation that became dominant last spring and forewarned the current explosion in variation, is one example. On the whole, the mutations in the receptor binding domain are more striking, if only because more parallels exist between them and the new variants threatening populations the world over.
Two of these substitutions, E484K and N501Y, are known to play a critical role in allowing variants that originated in the UK, South Africa, and Brazil to spread faster and further from host to host. The N501Y mutation, which substitutes one positively charged amino acid for one even more highly charged, increases the affinity of the spike for our receptors and was also seen in the London patient. The E484K mutation, on the other hand, is thought to decrease the potency of experimental therapies that rely on the antibodies of recovered Covid-19 patients to neutralize the virus, such as convalescent sera and monoclonal antibodies. E484K, in other words, probably assists SARS-CoV-2 in slipping past our immune systems, leaving us more vulnerable to disease.
Another substitution identified in the Boston patient, Q493K, was also found in another immunocompromised Covid-19 patient I’ll call the Italian patient, whose case I’ll discuss in a forthcoming piece. It appears that Q493K may facilitate escape from neutralization and increase binding affinities. The last two mutations in the receptor binding domain of note, T478K and S494P, have yet to arise in any of the variants currently circulating around the globe, but did appear in separate laboratory experiments conducted by scientists Jesse Bloom and Gideon Schreiber and could in future variants. Bloom’s study analyzed potential mutations for their ability to resist convalescent sera, while Schreiber’s looked at ones that increased binding affinity to the ACE2 receptor. Their work, along with the findings from the London and Boston patients, sheds light on the variations of SARS-CoV-2 that may blindside us further down the road.
Outside the N-terminal and receptor binding domains in the virus sequenced from the Boston patient are a couple more mutations worth mentioning, even though their purpose remains largely unknown. One substitution, I870V, falls between the fusion peptide and heptad repeat 1 regions of the spike. It is worth noting there are more changes in the viral genome, like ORF1a and ORF1b, where a deletion and three point mutations are located.
In the next installment of this series, I will discuss the Pittsburgh patient, yet another individual case of Covid-19 that might have implications for variants that may contribute to recurrent seasonal infections.
Originally published on Forbes (February 16, 2021)

