A New Variant In The Philippines
(Posted on Thursday, March 18, 2021)
A nurse prepares to administer the Sinovac COVID19 vaccine during a ceremonial vaccination program held inside a sports stadium in Marikina City, east of Manila, Philippines on 02 March 2021. According to the city health office, only 250 healthcare
NURPHOTO VIA GETTY IMAGES
Covid-19 has reached extraordinarily high levels in the Philippines. In early January, the country reported around 1,500 cases per day. Today, that rate is over 10,000, as seen below in figure 1. This resurgence of SARS-CoV-2 is coincident with the observation of a new Philippine variant: B.1.1.28.3. First identified in the Central Visayas region of the island nation, the variant is derived from the Brazilian B.1.1.28 strain and shares many mutations with the P.1 variant identified in Manaus in early January. However, this variant also has many characteristics unique to the Philippines. Here, we will analyze these unique and shared mutations and discuss their potential impact on the virus.
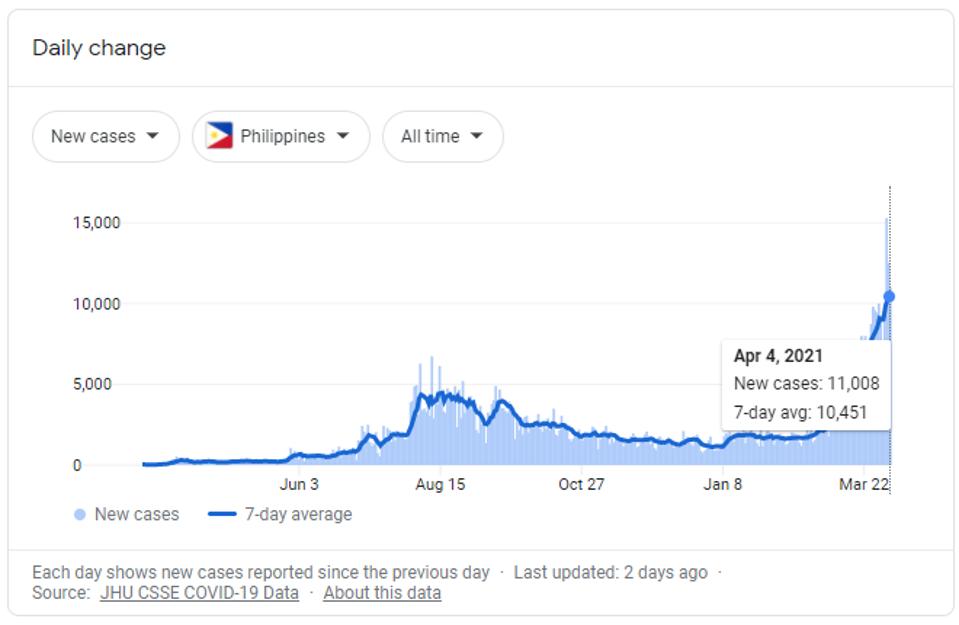
Seven-day rolling average of confirmed Covid-19 cases in the Philippines.
GOOGLE / JHU CSSE
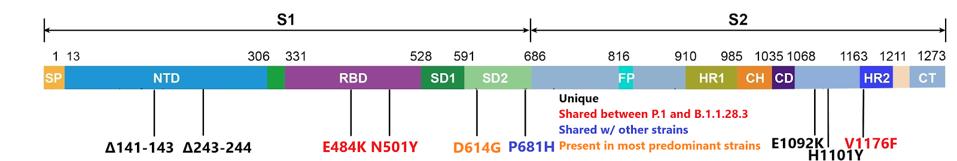
The B.1.1.28.3 variant contains the aspartic acid to glycine at position 614 (D614G) in the spike protein. This mutation was first spotted in the opening months of 2020 and later came to dominate nearly all strains of the virus in circulation today. The B.1.1.28.3 variant also contains spike mutations of glutamic acid to lysine at position 484 (E484K) and asparagine to tyrosine at position 501 (N501Y). The N501Y mutation is notable for its appearance in the widespread UK B.1.1.7 variant and the E484K mutation is similarly notable for its presence in the South African B.1.351 variant. Vaccine-elicited antibodies have been found to have reduced neutralization of pseudovirus containing these mutations.
In addition, the Philippine variant contains a number of other genome changes of note. Using the GISAID database, we will examine how many sequenced viruses share mutations with those found in the B.1.1.28.3 variant. Two sets of deletions from positions 141 to 143 and 243 to 244 in the N-terminal domain are found in only 72 of over 990,000 viruses sequenced on GISAID, most of which are concentrated in the Philippines. The deletions from positions 243 to 244 are notably present in a several months-long Covid-19 patient from Pittsburgh, indicating that these deletions may play a role in immune evasion as mutant viruses in long-Covid patients must adapt and evade the immune system to survive.
There are also two unique point mutations in the latter end of the spike sequence. Glutamic acid to lysine at position 1092 (E1092K) and histidine to tyrosine at position 1101 (H1101Y) are found in only 102 sequenced viruses. These two mutations are found predominantly in SARS-CoV-2 isolates from the Philippines. A study on SARS-CoV found this region susceptible to a series of neutralizing antibodies. Because much of the structure between SARS-CoV and SARS-CoV-2 is shared, it is possible that mutations to this site could additionally increase immune evasion, but further study on SARS-CoV-2 would be needed to confirm that hypothesis.
Despite what seems likely to derived from the B.1.1.28 lineage, the Philippine B.1.1.28.3 variant and the Brazilian P.1 variant differ substantially within the S protein and protein-coding regions of the virus, specifically ORF1a and several of the accessory proteins. The effect of each of the mutations outside of the spike protein deserved to be analyzed for the effect in cell culture and in animal models, alone and in combination, to assess their effects on the ability of the virus to replicate, transmit, and induce disease.
The figure below displays the full set of spike mutations.
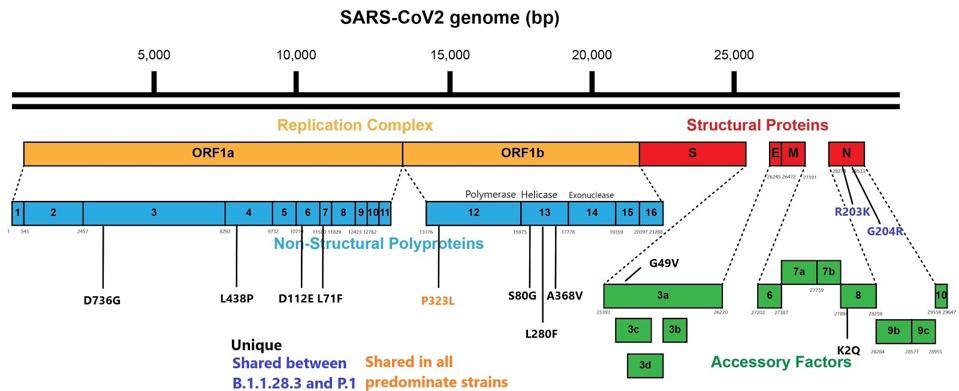
We also note that although many focus on the mutations in the spike protein, there should be an emphasis on the mutations in other proteins as determinants of transmission, virulence, and immune escape as well. While the spike protein is the mechanism controlling receptor binding and viral fusion with the host cell, the other proteins are significantly involved in pathogenesis, viral replication, and evasion from the immune system.
Some of the more notable mutations external to the spike protein include a change from arginine to lysine at position 203 (R203K) and glycine to arginine and position 204 (G204R) both in the nucleocapsid protein (N). The same mutations in N are present in other variants of concern including P.1, the UK B.1.1.7, the Danish B.1.1.298, and a cluster of 677 variants in the United States. The N protein is multifunctional. It stabilizes the virus particle, inhibits the interferon response to infection, and binds to genomic RNA. All three functions may contribute to infectivity and pathogenesis.
Three unique mutations of note are serine to glycine at position 80 (S80G), leucine to phenylalanine at position 280 (L280F), and alanine to valine at position 368 (A368V) in the nonstructural protein thirteen (NSP13) of ORF1a. NSP13 is an RNA helicase, critical to virus replication. The contribution, if any, to the efficiency of virus replication is worthy of further investigation. The figure below displays the full set of mutations external to the spike protein.
The B.1.1.28.3 variant is in the Philippines but is one of many unique variants popping up around the world. Each testifies to the ability of SARS-CoV-2 to adapt to new conditions of mitigation, prior immunity, and treatment. Continued vigilance coupled with government-mandated mitigation efforts coupled to rapid vaccination of entire populations may be the most effective means to blunt the potentially devastating impact of the onslaught of the variants.
We thank Dr. Maria Rosario S. Vergeire and the Philippine Department of Health for their insights in preparation of this updated report.
Read the full article on Forbes.
Originally published on March 18, 2021

