New Belgian Variant Illustrates The Versatility Of SARS-CoV-2 In Escaping Immune Suppression
(Posted on Thursday, April 1, 2021)

A single yellow tulip in a field of red
WANDERLUSTINGK.COM
There is now a new mutant to add to the global SARS-CoV-2 variant collection, this one in Belgium. This new Belgian variant (B.1.214), first detected in January by researchers at the University of Liege, although it is only a relatively small percentage of cases domestically and abroad, it appears to have the potential to become quite pervasive. It has mutations, not only in the spike protein as many others have, but also in other parts of the genome, all of which may contribute to increased transmission, virulence, and immune evasion.
Beginning within the spike protein, the B.1.214 variant carries six total mutations. The function of the spike protein is to seek out and bind to ACE2 receptors of human host cells in the nasopharynx, intestines, and other parts of the body. It allows the virus to bind and open an entry port to the cell to infect and spread within the host. This is a frequent site of mutations for past variants, some of which may cause increased virulence and infectivity. Although the B.1.214 variant lacks some of the most common mutations, such as E484K, N501Y, and others, it has unique mutations of concern. Of these mutations, we believe the following is the most potentially troubling.
An asparagine to lysine change at position 450 (N450K) lies in the receptor-binding domain, which binds directly to the ACE2. In other variants, this type of mutation may have a dual effect. First, a mutation to this mechanism could increase the affinity of the spike protein, leading to greater infectivity of the virus. Second, these types of mutations could abrogate the ability of neutralizing antibodies to bind, leading to a more immune-evasive virus. There are additional mutations in the spike protein that could be of similar concern. Shown below in figure one is the full wealth of mutations in the spike protein.
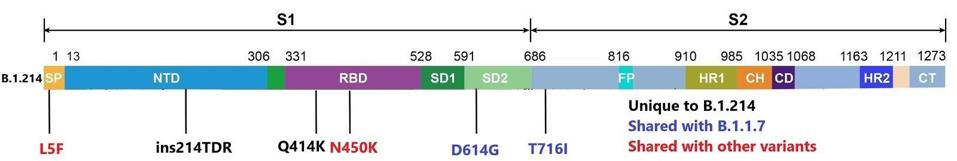
Mutations to the B.1.214 spike protein
Replicative Complex
Replication of the SARS-CoV-2 virus is complex. It involves the creation of specialized compartments to shield the virus from immune surveillance, as well as a unique and elaborate method of producing replicated viral proteins unique to this family of viruses. That requires the cooperation of at least sixteen proteins, the nonstructural proteins (NSP), many of which are multifunctional. Changes to the NSPs numbered one through sixteen can increase the ability of the virus to grow to rather high titers. A common characteristic among variants is a large number of mutations to this complex. These variants are also often noted to produce in high titers, likely leading to increased severity of disease. We note that the B.1.214 variant has eleven mutations, many of these in critical areas for facilitating virus replication.
Immune Modulators
The SARS-CoV-2 genome contains a set of proteins, as many as fourteen, which alter both the innate and the adaptive immune responses to its own advantage. Many pervasive SARS-CoV-2 variants, including the widespread United Kingdom (B.1.1.7) variant, have a number of mutations to these proteins as well. We note that the B.1.214 variant contains significant changes to the immune modulators, and the following are some of great concern.
In the nucleocapsid protein, the aspartic acid to leucine mutation at position 3 (D3L) lies in the signal peptide, which drives the protein through the membrane. The nucleocapsid protein provides stability to the virus, antagonizes viral RNA and interferon responses, and inhibits genome replication in the host cell, thus a mutation to this region could enhance these critical processes.
In the open-reading frame three (ORF3a) protein, there are a set of nine amino acid deletions from positions 20 to 28. ORF3a is additionally involved in antagonizing interferon responses and is a common site for mutations across SARS-CoV-2 variants. The B.1.214 variant is unusual in that ORF8, which is often heavily mutated or even eliminated in some variants, is fully intact. Shown below in figure 2 is the full wealth of mutations in B.1.214 external to the spike protein.
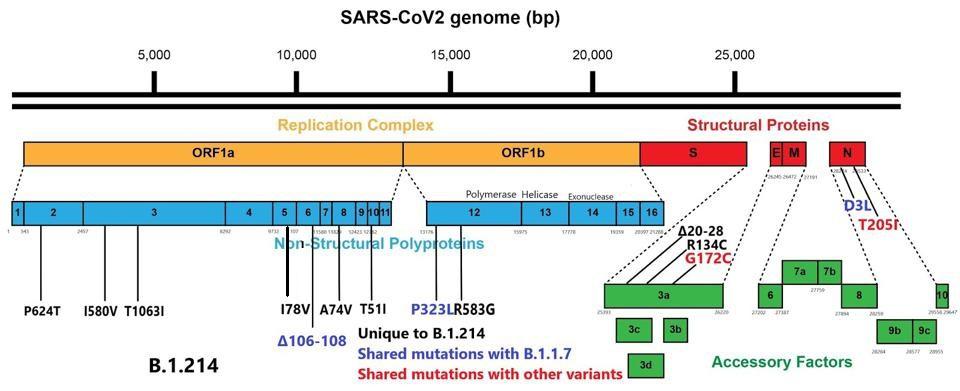
Mutations external to the spike protein
Origin
Notably, both the deletions in ORF3a and the D3L mutation in the nucleocapsid protein, among others, are also observed in the widespread B.1.1.7 variant. There are two possible explanations for this. Either they were derived entirely independently, or that they derive from a precursor variant. Given the significant number of mutations that these, and only these variants share, we speculate that both come from common stock. The lineage is shown in figure three below.
The B.1.214 variant shows a concerning combination of common mutations that have caused trouble with previous widespread variants, as well as unique mutations which may only add to the dangerous recipe. We suggest it receive continued monitoring and research as it could be a driver of new surges in cases yet to come.
One current speculation is that SARS-CoV-2 has a limited set of mutations, which it may have exhausted. If this is true, it would be far more susceptible to second and third-generation vaccines. We find this conclusion unlikely. As you can see from the B.1.214 variants and others we have previously discussed, there is a wide array of variants that spread widely, despite lacking the popular N501Y and E484K mutations, such as the California and Washington DC variants.
Additionally, the cold-causing coronaviruses recur annually and studies show the spike protein can vary 4% between cold seasons and the receptor-binding domain specifically can vary up to 17%. We estimate this is a lower limit on the mutative capabilities of SARS-CoV-2, in other words, we have not begun to see the virtuosity of this virus in its wealth of mutation and immune evasiveness.
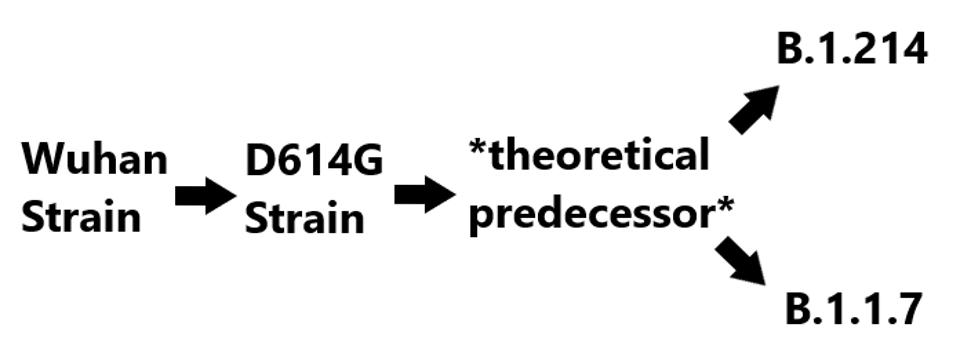
Proposed B.1.214 lineage
(FIGURE 3) The theoretical lineage of B.1.1.7 and B.1.214. The theoretical predecessor contains the shared mutations between B.1.214 and B.1.1.7, those being 5’ UTR: C241U, NSP6: Δ106-108, NSP12: P323L, S: D614G and T716I, N: D3L. Unique mutations in B.1.214 are NSP3: I580V and T1063I, NSP5: I78V, NSP8: A74V, NSP10: T51I, NSP12: R583G, S: L5F, ins214TDR, Q414K, and N450K, ORF3a: Δ20-28, R134C, and G172C, N: T205I. Unique mutations in B.1.1.7 are NSP3: T183I, A890D, and I1412T, NSP13: T239I and K460R, S: Δ69-70, Δ144, N501Y, A570D, P681H, S982A, D1118H, ORF8: Q27stop, R52I, and Y73C, N: R203K, G204R, and S235F.
Read the full article on Forbes,
Originally published on April 1, 2021.

