An Indian SARS-CoV-2 Variant Lands In California. More Danger Ahead?
(Posted on Monday, April 12, 2021)

Gateway to Mumbai
A.SAVIN, WIKIMEDIA COMMONS
Yet another variant of SARS-CoV-2 has arrived on our shores from afar. A recent survey of the viral sequences infecting Californians turned up six independent isolates of a variant that is now circulating widely in India. Over the past two months, infection rates in India have exploded.
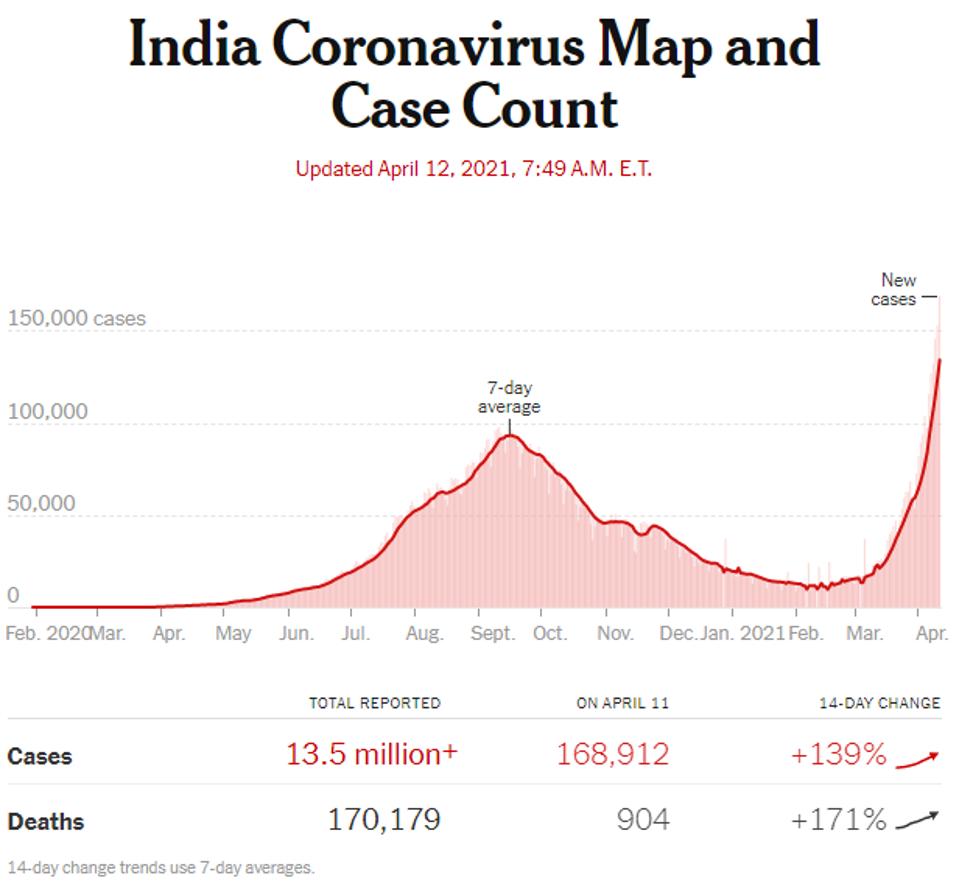
Confirmed Covid-19 cases in India
THE NEW YORK TIMES
On April 9th, 145,384 India reported 145,382 new infections, a tenfold increase over the 11,012 new infections reported on February 9, just two months ago. The Indian variant designated B.1.617 is likely, at least in part, to be driving the current wave. The new strain has been detected in at least 5 Indian states, including Maharashtra, Delhi, and Punjab, which have all contributed to the surge of cases in the country, in addition to the B.1.1.7 (UK), B.1.351 (South Africa), and P.1 (Brazil) variants which are also circulating India. It is unknown the extent to which this strain has spread, but of the 2,844 sequences submitted to the GISAID database from India since March 1, 162 sequences have been classified as B.1.617, or about 6%. This percentage is likely to grow. This is all the more reason for concern now that this variant has arrived here in the United States.
Detailed analysis of the genome and proteins of B.1.617 reveal it arose independently in India. It differs substantially from the B.1.1.7, P.1, B.1.351, B.1.427/9 (California), and B.1.526 (New York) variants currently circulating in the United States. All of the 15 amino acid changes of B.1.617 differ from those of all other variants of concern when compared to the globally dominant D614G strain designated B.1 with two notable exceptions.
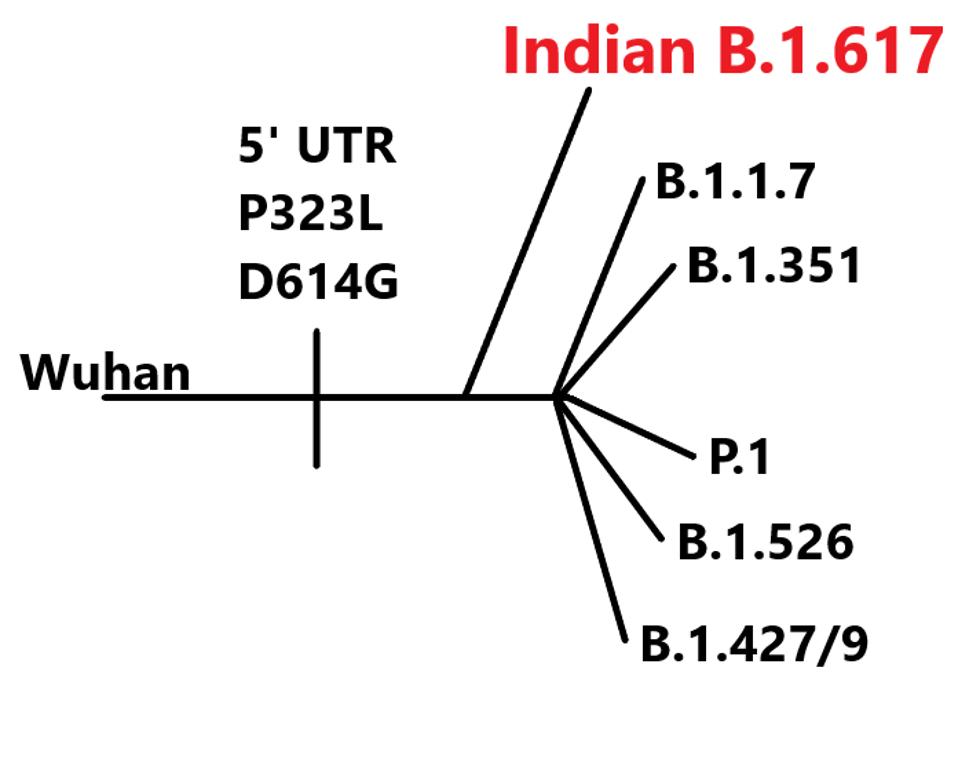
Lineage of B.1.617 variant
The exceptions are in a region of the spike protein, the receptor-binding domain (RBD), that is critical both for binding to the ACE2 receptor and antibody neutralization.
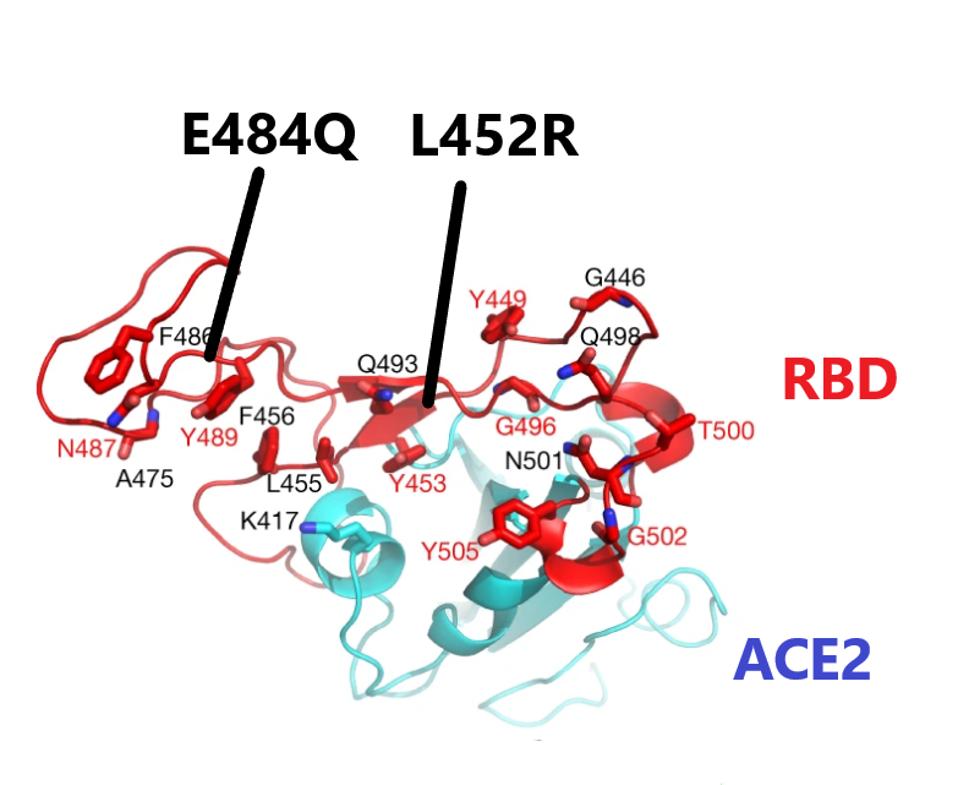
The first of these is a leucine to arginine substitution at amino acid 452 (L452R). This is the same change found in the California variant (B.1.427/429). Laboratory experiments demonstrate that this change both increases the affinity of the spike for the receptor and decreases antibody recognition, including recognition by antibodies present in convalescent serum and as well as some clinically important neutralizing monoclonal antibodies.
The second mutation of interest occurs at amino acid 484. Many of the variants of concern, including the B.1.351, P.1, and B.1.526 variants, substitute glutamic acid for lysine (E484K), which is a substitution of a negatively charged for a positively charged amino acid. This change reduces neutralization by convalescent antisera and binding of some monoclonal antibodies and increases the affinity for ACE2.
The Indian variant B.1.617 is also mutated at position 484. However, the 484 mutation is different. The glutamic acid is substituted by the polar uncharged amino acid glutamine (E484Q). Laboratory experiments confirm that this change also confers increased ACE2 binding and immune evasion properties. The combination of the two, sometimes called the double mutant, goes a long way toward explaining the increased transmission of the India variant in its home country, and highlights the danger now that it is established here in the United States.
Two of the five remaining mutations in the spike protein are located in the N-terminal domain of the S1 protein, a region that is extensively mutated in other variants of concern. These two mutations are unique to the Indian variant.
The third mutant in S1, the substitution of proline for arginine (P681R), is located near the cleavage site between S1 and S2. This change may increase the infectivity of the virus particles by facilitating cleavage of the S precursor protein to the active S1/S2 configuration. The contribution of the two remaining amino acid substitutions in the S2 portions is also unique to the Indian variant. The full extent of spike mutations in the B.1.617 variant can be found in the figure below.
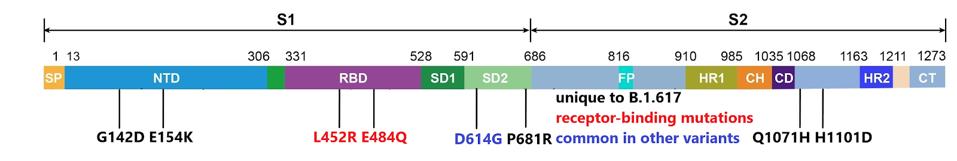
Most of the variants of concern also contain an extensive set of mutations in the structural proteins, the replication enzymes, and the accessory proteins. B.1.617 is no exception. B.1.617 carries one mutation in each of the replication enzymes, NSP3, NSP6, NSP13, NSP15, NSP16 as well as one mutation in the accessory proteins orf3a, orf6, and orf7a. As before, these changes are unique to the Indian variant. All these mutations are illustrated in the following figure.
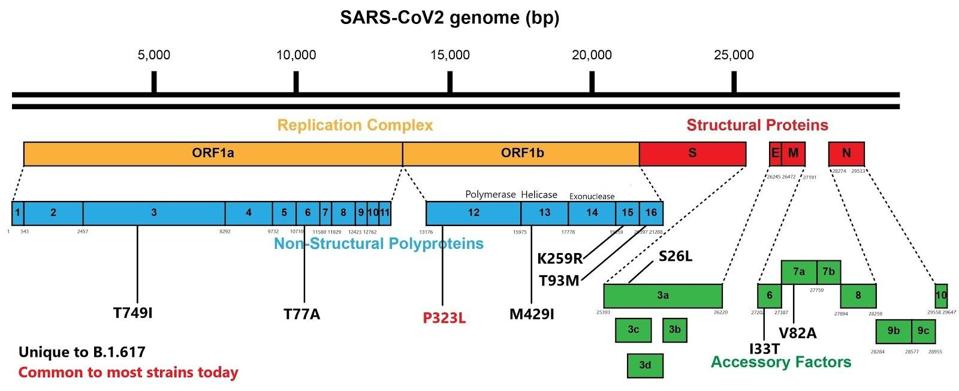
B.1.617 genome with mutations
It is still early days in understanding the importance of the arrival of the Indian variant in the United States. The B.1.617 variant has all the hallmarks of a very dangerous virus. If indeed it is this variant that is driving the unprecedented exponential increase in infections in India, the potential for trouble here in the United States is real and immediate. We must do all that is possible to identify its spread and to contain it.
We thank Dr. Benjamin Pinsky and Dr. Malaya Kumar Sahoo for their insights in preparation for this report.
April 16th Update: Seventy-seven new cases of the B.1.617 variant have been discovered in the United Kingdom, according to a report by the Financial Times. The virus is rapidly spreading in India and “confirmed cases [in the UK] are growing at a similar rate to B.1.1.7 at the same stage of its emergence.” This variant must continue to be monitored as it appears to have rapid-spread capabilities like many variants of concern.
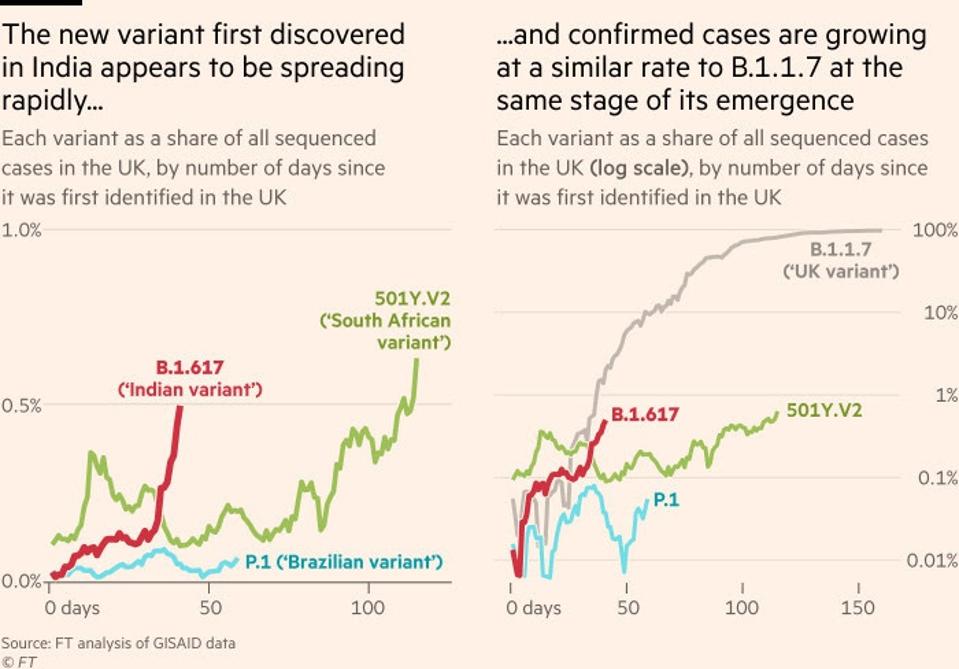
The rapid rise of B.1.617
THE FINANCIAL TIMES
A number of people have minimized the variants in terms of their potential to spread, reinfect, and avoid vaccination. As I look around the world, I find variants are increasing their cause for concern in all respects. This discovery only reinforces the urgency to dramatically increase domestic and international surveillance of disease to prepare us for what appears to be a continuous onslaught of SARS-CoV-2 variants.
Read the full article on Forbes.
Originally published April 12, 2021.

