A Newly Discovered Antibody Neutralizes Many Variants By Locking The Receptor-Binding Domain In A Closed Position
(Posted on Tuesday, May 11, 2021)
As SARS-CoV-2 variants grow in type and frequency, Covid-19 researchers are on the hunt for parts of the virus that remain consistent across variants in order to create Covid-19 treatments that work for multiple strains of the virus. This is the fourth in a series discussing these potential Achilles’ heels for Covid-19. Read more from this series in parts one, two, and three.
Antibodies
GETTY IMAGES / CHRISTOPH BURGSTEDT
Cocktails of monoclonal antibodies are valuable therapies for the prevention and treatment of Covid-19. However, their utility has been compromised by variants of SARS-CoV-2 that evade one or more of the antibodies in the mix.
A new paper by Scheid et al. describes a new monoclonal antibody—BG10-19—that has a valuable set of characteristics. It is highly potent and active against a broad range of variants. BG10-19 and other antibodies analyzed in the study bind to the receptor-binding domain of the spike protein. The receptor-binding domain can be either open or closed. It must be in the open configuration to bind the ACE2 receptor to initiate infection.

FIGURE 1: Receptor-binding domain open and closed
STANFORD UNIVERSITY
The BG10-19 antibody acts by bonding tightly to the top of the trimeric receptor and locking all three receptor-binding domains in the closed configuration (Figure 2).
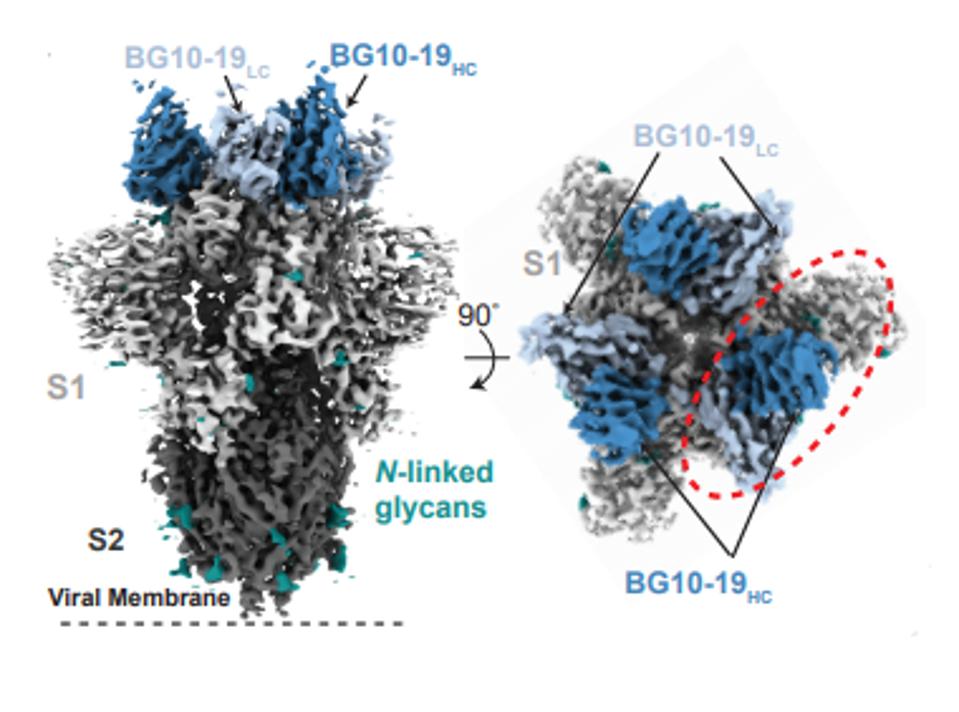
FIGURE 2: BG10-19 binding to the RBD
SCHEID ET AL.
The special properties of BG10-19 derive, in part, its ability to bind to two adjacent receptor-binding domains simultaneously. In figure 3, the majority of the interactions are pictured with the receptor-binding site “RBD-A,” while binding simultaneously to the adjacent receptor-binding domain of the trimer designated as “RBD-B.” BG10-19 binds to two N-linked glycans on RBD-A as well.

FIGURE 3: BG10-19 bridging across RBDs
SCHEID ET AL.
This versatile antibody was discovered through a selective process of elimination. B cells from 14 patients who had recovered from mild cases of Covid-19 were isolated. The researchers selected 6,113 B cells that either bound to the SARS-CoV-2 S-protein or to the receptor-binding domain. They narrowed the search to B cells originating from four of the 14 patients who had the highest concentration of neutralizing antibodies against SARS-CoV-2 lentivirus pseudotyped with the B.1 spike protein, derived from the wild-type Wuhan strain carrying the D614G spike mutation.
The FAB fragments of 92 monoclonal antibodies isolated from the four patients were added to an IgG1 framework. They then screened the 92 antibodies for polyreactivity, nonspecific binding to unrelated antigens, testing for binding to single-stranded DNA, double-stranded DNA, insulin, bacterial lipopolysaccharide, and streptavidin-APC. They found that 11 of the 92 monoclonal antibodies showed reactivity against two or more of the listed antigens. These were eliminated from future consideration.
The search narrowed further by selecting those that bound the SARS-CoV-2 spike protein and the receptor-binding domain. Forty-two of the antibodies bound to both, nine bound exclusively to the spike protein, and five bound exclusively to the receptor-binding domain. Next, they screened the antibodies for neutralization capability against both SARS-CoV-2 pseudotypes and live virus (the B.1 variant of SARS-CoV-2). Twenty-seven of the 92 showed neutralizing activity with IC50 values ranging from 3 ng/ml to 25 µg/ml.
They narrowed their search to six potential antibody candidates based on neutralization potency. BG10-19 was selected for further study as it was one of the most potent neutralizing antibodies and capable of neutralizing two variants of concern, the B.1.1.7 variant first isolated in the UK and the B.1.351 variant that was discovered in South Africa (Figure 4).

FIGURE 4: BG10-19 neutralization of the wild type and variants
SCHEID ET AL.
The set of antibodies was also tested for its ability to bind to spike proteins, each of which carried a single mutation on the spike found in many variants of concern. The heat map of figure 5 summarizes the decrease in binding affinity of each antibody to each variant spike protein, the deeper the red coloration the greater the decrease in binding. Two of the tested antibodies, BG10-19 and BG1-28, are largely unaffected by any single mutation and bind to both SARS-1 and the WIV1 spike proteins. Sequence conservation at the BG10-19 epitope explains the cross-neutralization against SARS-1, as 23 of 29 residues are conserved between SARS-1 and SARS-CoV-2 RBDs. Notably, despite strong binding to the WIV1 spike protein, there is little neutralizing activity, despite a highly parallel amino acid sequence to SARS-1. Again, BG10-19 was selected for further study as it binds and neutralizes SARS-CoV-2 much with a much lower IC50 than does BG-1-28 (Figure 5).
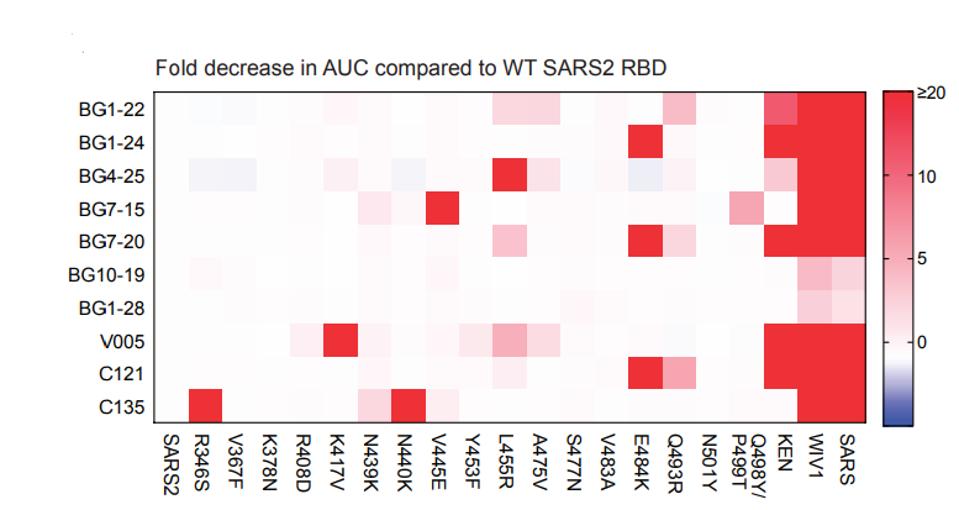
FIGURE 5: Heat map of mutations that reduce neutralizing capabilities of different antibodies.
SCHEID ET AL.
This result is somewhat surprising as at least five of the spike proteins tested alter amino acids well within the binding footprint of BG10-19, specifically the mutations R433S, V367F, N439K, N440K, and V445E. Two of the naturally occurring mutations, N439K and V445E, do reduce binding affinity modestly between 1 and 5 fold.
It is notable that the binding site of BG10-19 does not overlap with the residues of the receptor-binding domain that interact directly with the ACE2 receptor. Additionally, the binding site of another broadly neutralizing monoclonal antibody selected for clinical development S309 overlaps that of BG10-19 (Figure 6).
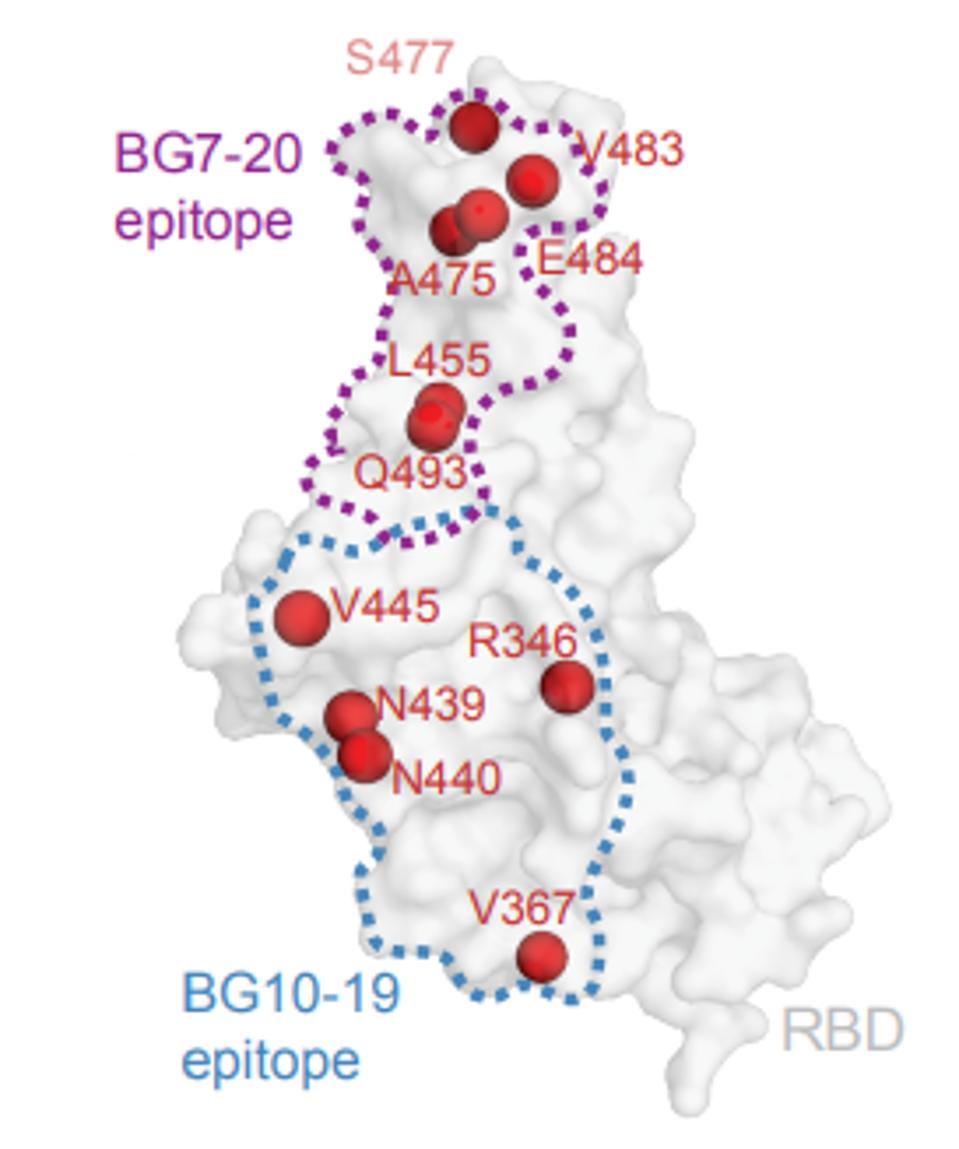
FIGURE 6: Antibody footprint of BG10-19
SCHEID ET AL.
This is not to say there are no escape variants that reduce binding and neutralization to the BG10-19 antibody. In fact, they found two mutations that reduce BG10-19’s neutralizing capability: G339R and L441P. The G339R mutation reduces neutralization as much as ten-fold and the L441P mutation reduces neutralization about one hundred-fold. While these mutations have not yet been sequenced and deposited into the GISAID mutation database, there have been other mutations at both positions. There have been 238 sequenced mutations at position 339, including changes to aspartic acid, serine, valine, as well as some deletions. Additionally, there have been 101 sequenced mutations at position 441, including changes to isoleucine, phenylalanine, arginine, as well as some deletions. While these are not the mutations that inhibit neutralization, their existence indicates that mutations to these positions are tolerated.
All of the antibodies discussed in this paper are very interesting in terms of their potential use. They could be used in various combinations. We suggest the BG10-19 antibody be combined with the S309 and S2H97 antibodies described by Starr et al. to create a cocktail that recognizes conserved epitopes and that is both potent and resistant to naturally arising variants.
Read the full article on Forbes.
Originally published on May 11, 2021.

