How SARS-CoV-2 Evades And Suppresses The Immune System (Part 13)
(Posted on Wednesday, October 6, 2021)
This is the thirteenth article in a series called “How SARS-CoV-2 Evades And Suppresses The Immune System,” which will explore an under-appreciated but highly significant aspect of SARS-CoV-2 replication. The ability of SARS-CoV-2 to delay, evade, and suppress the immune system has myriad implications for drugs, vaccines, and other aspects of our pandemic response. The first set of pieces in this series are intended for a general audience; the second set, for the medical community; and the third and final set, for biomedical researchers looking for a deeper understanding of variants, how they’re generated, and what we might do to control them. Read parts 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, and 12.

As a fullback James Develin normally ran the ball. Here he is shown doing double duty preparing for a forward pass.
NBC SPORTS HTTPS://WWW.NBCSPORTS.COM/BOSTON/NEW-ENGLAND-PATRIOTS/JAMES-DEVELIN-STAYS-TOP-TIGHT-END-TECHNIQUES-CASE-HES-NEXT-MAN-NEW-ENGLAND-PATRIOTS
Double Duty by the Membrane Protein M
In previous installments of this series I discussed how different SARS-CoV-2 viral proteins suppress innate immunity by inhibiting production of interferons and induction of interferon-stimulated genes.
Recall that one of the ways the virus suppresses innate immunity is by interfering with multiple steps along key interferon signaling pathways (see Figure 1). In Part 11 and Part 12 of this series I described how SARS-CoV-2 inhibits the retinoic acid-inducible gene-I (RIG-I) pathway through the nonstructural proteins NSP1, NSP6, and NSP13; the open reading frames Orf3a, Orf6, Orf7a, and Orf7b; and the membrane (M) protein. In those pieces I mainly examined the role played by the nonstructural proteins and open reading frames. Today we turn our attention to the last remaining piece of the puzzle, the M protein.

Figure 1. SARS-CoV-2 and its effect on interferon. RIG-1 (retinoic acid-inducible gene 1) is inhibited by NSP1; MAVS (mitochondrial antiviral signaling protein); IKKε (inhibitor of nuclear factor kappa-B kinase subunit epsilon) is inhibited by NSP13;
ACCESS HEALTH INTERNATIONAL
The primary function of RIG-I is to recognize and bind to double stranded RNA, a key intermediate in SARS-CoV-2 replication and transcription. Upon binding to double stranded RNA, RIG-I activates mitochondrial antiviral adaptor protein (MAVS), which in turn sets off a cascade of downstream factors: k-B kinase ε (IKKε), TANK binding kinase 1 (TBK1), and interferon regulatory factor 3 (IRF3). If phosphorylated, interferon responsive factor 3 translocates to the nucleus and initiates transcription of type-I interferons.
A study published in October 2020 found that overexpression of the M protein in cells infected with SARS-CoV-2 led to enhanced viral replication and inhibition of the innate immune response. More specifically, the researchers found that the M protein directly targets MAVS, the protein that recruits downstream antiviral genes into initiating interferon transcription. In antagonizing MAVS, the M protein obstructs the formation of the polyprotein complex that facilitates phosphorylation and dimerization of interferon responsive factor 3. It does this by binding to MAVS directly.
Another study, published more recently in May 2021, took a closer look at this mechanism to see whether, in addition to inhibiting MAVS, the M protein interferes with RIG-I, MDA5, or TBK1. The authors of the study identified TBK1 as another target of the M protein, a kinase critical to the phosphorylation of interferon responsive factor 3 (see Figure 2). Evidently the SARS-CoV-2 M protein inhibits TBK1 by introducing a post-translational modification, in this case a ubiquitin, that facilitates its degradation, effectively terminating downstream signaling.
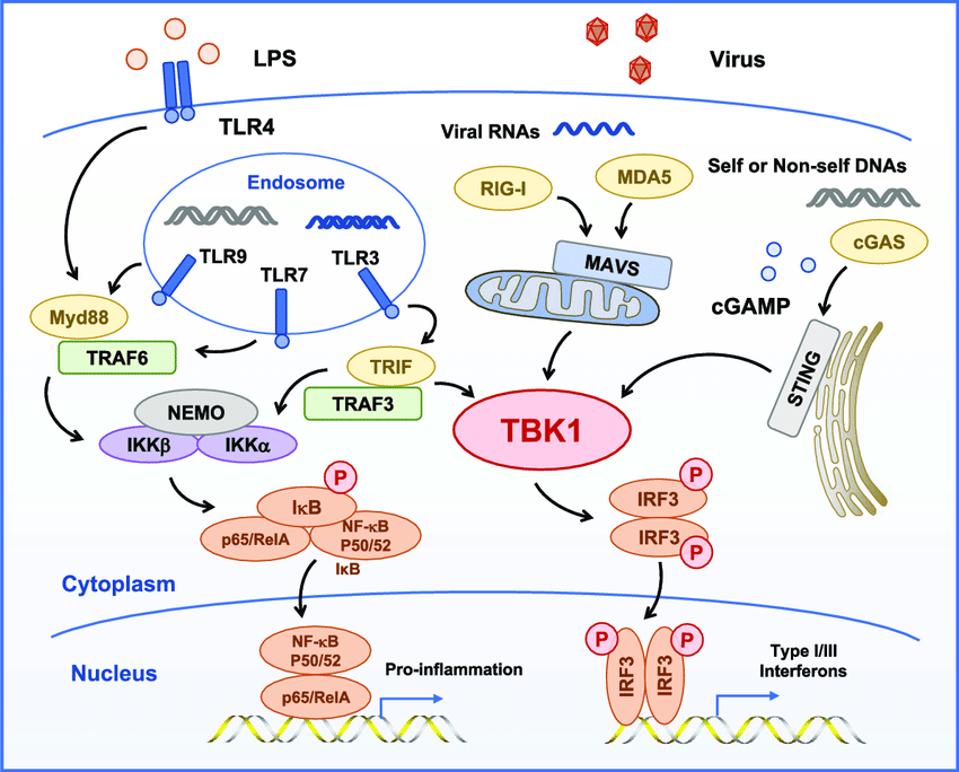
Figure 2. TBK1 is the central kinase of the MAVS, STING, and TRIF signaling complexes TBK1 directly phosphorylates of IRF3, MAVS, STING, and TRIF and is required for the assembly, activation, and IRF3 selectivity of these signalosomes.
TBK1, A CENTRAL KINASE IN INNATE IMMUNE SENSING OF NUCLEIC ACIDS AND BEYOND HTTPS://WWW.RESEARCHGATE.NET/FIGURE/TBK1-IS-THE-CENTRAL-KINASE-OF-THE-MAVS-STING-AND-TRIF-SIGNALING-COMPLEXES-TBK1-DIRECTLY_FIG3_341710443
Figure 2 illustrates two interrelated but independent pathways that stimulate responses to viral infections. RIG-I recognizes uncapped cytoplasmic RNAs and short, double-stranded RNA, whereas MDA5 recognizes longer double-stranded RNAs. Both trigger production of interferons, which in turn stimulate many genes and proteins that interfere at multiple stages with virus replication. On the right hand side of the figure, note that STING bypasses the M protein and many other sites along the RIG-I pathway by directly stimulating TBK1, which in turn stimulates interferon responsive factor 3 (IRF3). In a recent story, I described how dinucleotide activation of STING has a potent anti-SARS-CoV-2 effect in hamster models. This observation provides independent support of the critical role of viral proteins in innate immune suppression and virus replication and pathogenesis. On the left hand side lies a separate but interacting pathway that begins in the endosome. I’ll describe the role of the proinflammatory pathway in severe Covid-19 in my next piece.
In conclusion, evidence has established M to be yet another of the viral proteins that SARS-CoV-2 uses to suppress innate immunity, buying time to enter, replicate, and exit before the innate immune system kicks in.
Originally published on Forbes (September 22, 2021)

