A Single Point Mutation In The Nucleocapsid Protein Increases Covid Virus Infectivity By More Than 100 Times
(Posted on Tuesday, November 9, 2021)
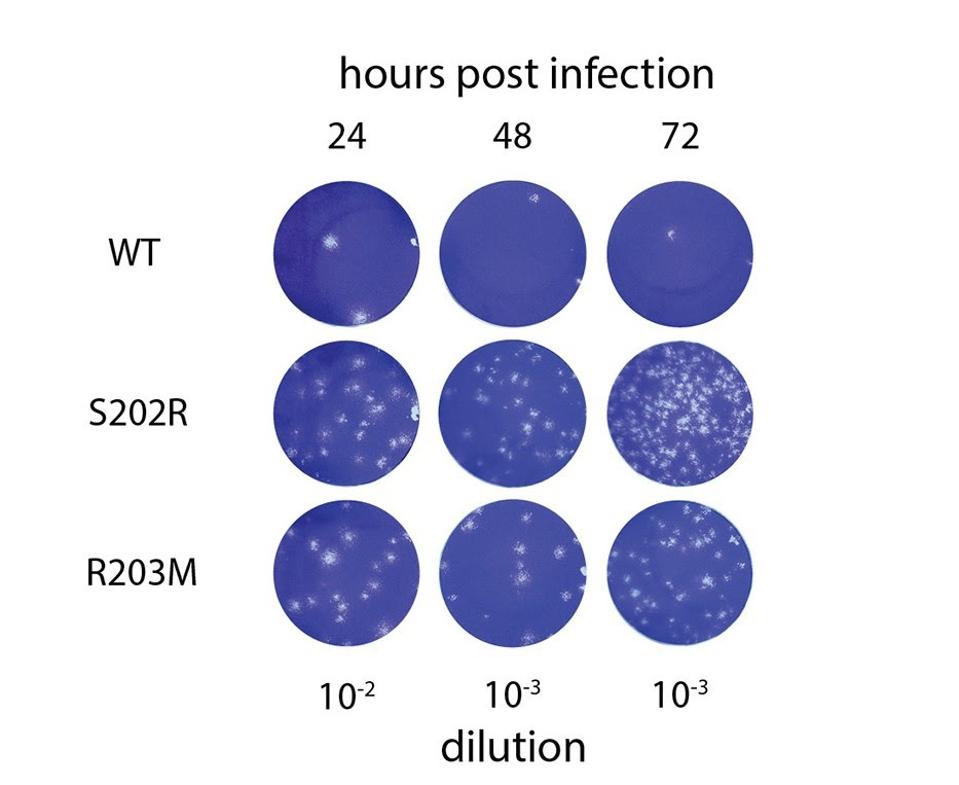
Increased plaque forming capability of N protein mutant viruses carrying either S202R or R203M mutations
SYED ET AL.
A recent paper by Syed et al. demonstrates that mutations in the Nucleocapsid (N) protein, one of the four structural proteins in SARS-CoV-2 in addition to the Spike protein, plays a critical role in the increased transmission and possibly virulence of the Delta variant. Previous studies of increased transmission and other virological properties of SARS-CoV-2 variants focus almost exclusively on the Spike (S) protein. This work emphasizes that it is of equal importance to look at the effects of mutations throughout the entire viral genome.
SARS-CoV-2 Virus-like Particle and Identification of the Packaging Sequence
To study the effects of structural protein mutations on the infectivity of SARS-CoV-2, the authors created a new experimental system which they call SARS-CoV-2 virus-like particles (SC2-VLPs). These are particles in which the structural proteins, N, S, the Envelope (E), and Membrane (M), exist as heterologous RNA which contains the SARS-CoV-2 packaging signal.
Creating the particles first required identification of the SARS-CoV-2 cis-acting packaging sequence. Packaging is the incorporation of RNA into host cell nuclei for replication. Through analysis of previous research on SARS-CoV and Murine Hepatitis Virus, they were able to refine the packaging signal down to a short sequence, specifically nucleotide positions 20080-21171.
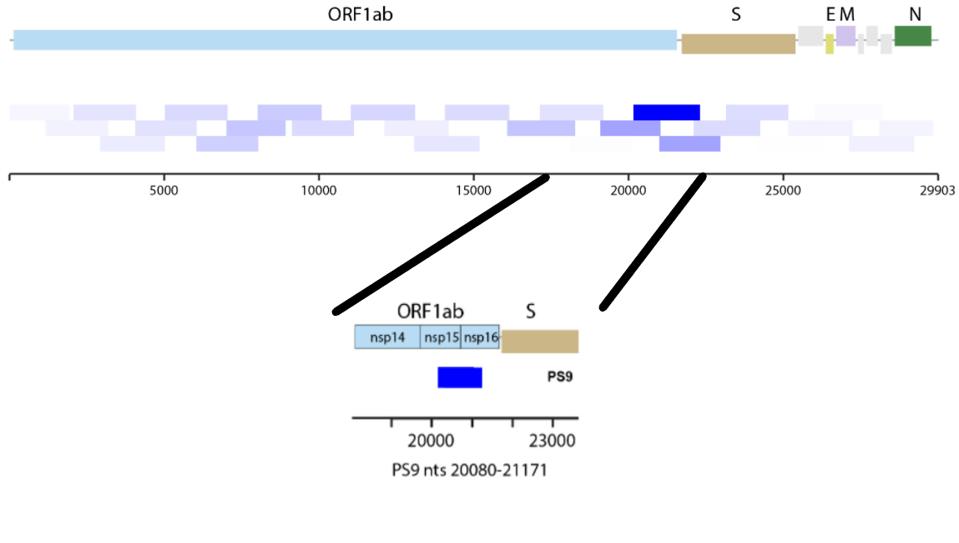
FIGURE 1: Visualization of the region where the identified packaging signal (PS9) is located in the viral genome.
SYED ET AL.
Isolation of this sequence enabled the fixed packaging of heterologous RNA into the virus-like particles.
FIGURE 2: Schematic representation of SARS-CoV-2 virus-like particles alongside a SARS-CoV-2 virus.
SYED ET AL.
Effects of Mutations on the S Protein
The authors used the SC2-VLPs first to examine the transmissibility of key S mutations to the Wuhan wildtype. Exposing the SC2-VLPs to 293T ACE2-imitating cells, the regulation of luciferase activity, which is positively associated with the more efficient assembly of the virus-like particles and subsequently stronger mRNA delivery. Notably, they found that none of the generated SC2-VLPs generated with S mutant genes, including four with the combined mutations found in the Alpha, Beta, Gamma, or Epsilon variants, increased luciferase expression to a significant extent. This observation that the S protein mutations have little to no effect on the ability of the virus to enter ACE2-positive cells differs somewhat from previous studies showing that Spike protein mutations confer some entry advantage.
Effects of Mutations on the N protein.
As they did for mutations in S protein, the authors generated SC2-VLPs which encoded major N protein mutations found in many naturally-occurring SARS-CoV-2 variants. The researchers note that all variants of concern contain at least one amino acid mutation between positions 199 to 205 of the N protein, indicating great significance in some form. This specific region is the linker domain between the N-terminal and C-terminal domains. The major mutations in this region are P199L, S202R, R203K/M, G204R, and T205I, at least one of which is found in every major natural variant.
Again exposing the SC2-VLPs to 293T ACE2-imitating cells, they found that the natural mutations, such as S199L, S202R, and R203K/M, increase luciferase activity, and thus transmissibility, by four to seven-fold. This observation suggests that the N protein, itself, conveys a replication advantage at least to SC2-VLPs.
To confirm their observations of greater luciferase and mRNA delivery activity in SARS-CoV-2 virus-like particles, the researchers then reverse-engineered SARS-CoV-2 cells in vitro using the S202R and R203M N protein mutations, which are commonly found in natural variants like Delta, with an original Wuhan strain genome. Infecting A549-ACE2 cells with wild-type virus, virus with S202R, and virus with R203M, they measured the RNA content and compared. PCR analysis indicated 45-fold higher RNA content and 166-fold higher infectious titers in viruses with S202R as compared to the wildtype. Additionally, R203M viruses posted 23-fold higher RNA content and 51-fold higher infectious titers. In other words, viruses with the mutant N protein bolstered infectiousness by around 50-fold.
Infectivity of Viruses Carrying N Protein Mutations
Further analysis of the infected cells used in the previous experiment 24, 48, and 72 hours post-infection found that infectious titers were 166-fold higher in viruses with S202R as compared to the wildtype. Additionally, R203M viruses posted 51-fold higher infectious titers. These experiments demonstrate that the N protein is a major contributor to increased replication of natural variants, specifically the Delta variant, which carries R203M, and possibly others.
The increased replication efficiency due to N protein mutations is significant, however, it only represents a fraction of the infectious capabilities of viral variants. The Delta variant, for instance, produces roughly 1000-fold more virus in nasal-pharynx secretions than the Wuhan virus.
We speculate that the other viral proteins and cis-acting packaging sequences may contribute to the additional 950-fold increase in replication efficiency. The Delta variant has 15 nonsynonymous mutations outside of the S and N proteins. These include the replicative-transcription complex (NSP1-16), the other structural proteins (E and M), and the regulatory Orf proteins (Orf3a, Orf7a, Orf7b, etc). Amongst these mutations are two mutations that are common to all natural variants along with D614G in the S protein, part of what we refer to as the Triad. These are P323L in the NSP12 polymerase and C241U in the 5’ untranslated region.
FIGURE 3: C241U in the 5’ untranslated region and P323L in the NSP12 polymerase.
ACCESS HEALTH INTERNATIONAL
This is a common theme throughout all natural variants of SARS-CoV-2. Below is a figure displaying the mutational capability of the virus.
FIGURE 4: SARS-CoV-2 genome. Bars indicate mutations in non-Spike protein present in at least 1% of SARS-CoV-2 sequences in GISAID sequence database.
ACCESS HEALTH INTERNATIONAL
Conclusion
A single point mutation in the N protein can increase the infectivity of the SARS-CoV-2 virus by 50-fold. Additionally, experimentation via SC2-VLPs is an elegant medium to potentially analyze virological impacts by mutations on other SARS-CoV-2. We hope that similar elegant methods can be applied to investigate the contributions of the other structural proteins, the replicative transcription complex, and the regulatory Orf proteins. Analysis of mutations in these proteins and in the genomic RNA sequence itself, which are plentiful throughout natural SARS-CoV-2 variants, may yield results akin to those found in the N protein.

