How Omicron Evades Natural Immunity, Vaccination, And Monoclonal Antibody Treatments
(Posted on Friday, December 17, 2021)
This is the third in our series on the Omicron variant. Find parts one and two here.
In a few short weeks, the COVID-19 virus variant Omicron has spread around the world. The incidence of new infections is rising rapidly, even in well-vaccinated populations and those previously infected by earlier variants of SARS-CoV-2. The epidemiologic evidence strongly points to a variant that is resistant to most if not all extant vaccines, and possibly many monoclonal antibodies treatments. Here we explore these concerns. This is the third in our series that outlines what we know about Omicron. We summarize the finding of recent experiments by Cameroni et al. in a bioRxiv preprint from December 14th.
Omicron ACE2 Binding
The first question asked was how tightly the Omicron Spike (S) protein binds to the ACE2 receptor. The data is summarized in Figure 1. Cameroni et al. show that Omicron’s affinity for the ACE2 receptor is 2.5 times as great as that of the S protein from the original Wuhan isolate. Omicron binds to the receptor as well as the Beta variant, but not as well as Alpha, which binds ACE2 almost six times more tightly. The N501Y mutation in Omicron is universally observed to increase affinity roughly 6-fold, yet other mutations in key sites like K417N, Q493R, and G496S were shown by deep mutational scanning to decrease affinity. Increased affinity for the receptor may account, in part, for increased transmissibility, but that is clearly not the whole story as Omicron is much more transmissible than any previously isolates, including Alpha, Beta, Gamma, and Delta.
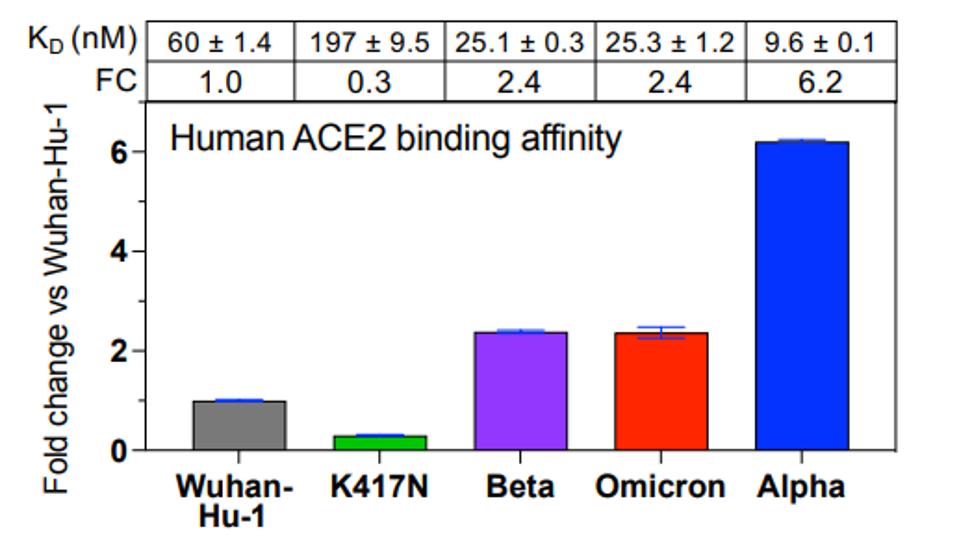
FIGURE 1: ACE2 binding of different SARS-CoV-2 variants.
CAMERONI ET AL.
Omicron Host Range
They next examined the Omicron host range. The ACE2 receptors of animals differ slightly in amino acid sequences in those regions of the protein which directly interact with the receptor-binding domain of the S protein. To assess the possibility that the extensive variation in the receptor-binding domain might affect the virus host range, the ability of the S protein to bind the ACE2 from three species—mink, pangolin, and mouse—was examined. The data from Cameroni et al. shows that the Omicron S protein binds to the Mouse ACE2, but not to those of mink or pangolin. There was no cross-species binding detected from the Wuhan, Alpha, or Beta isolates. They attribute this cross-affinity to the Q493R mutation in the Omicron receptor-binding domain, which is similar to the Q493K mutation isolated from mouse-derived SARS-CoV-2.
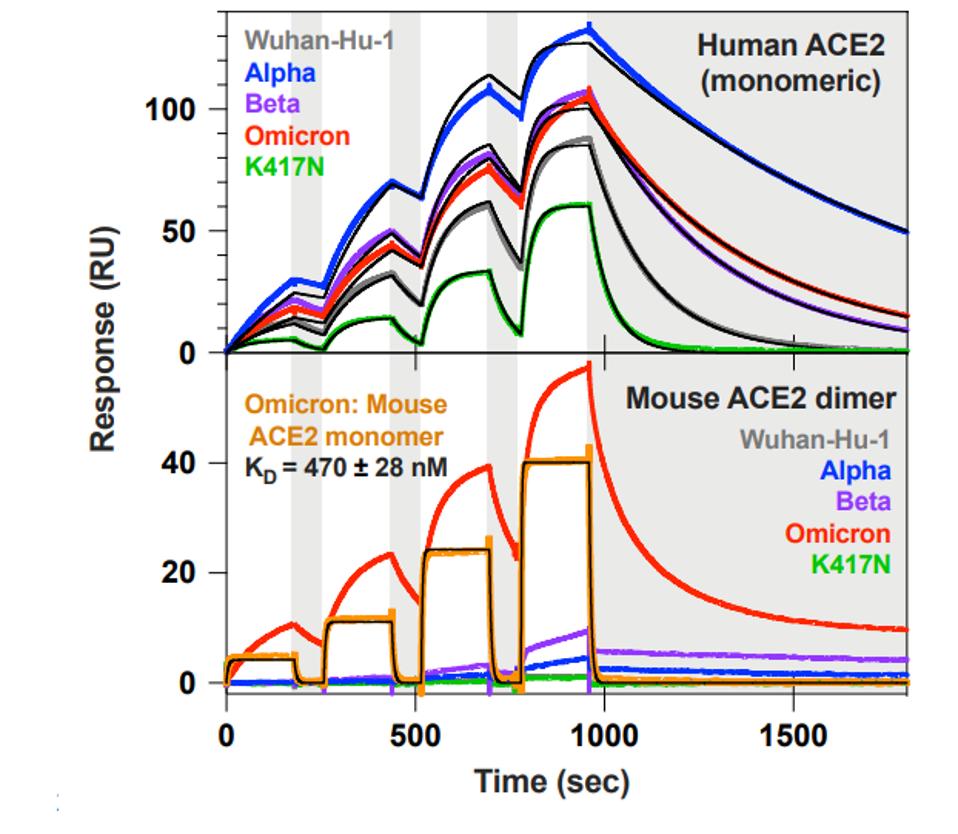
FIGURE 2: ACE2 binding kinetics of different SARS-CoV-2 variants to a mouse and human receptor.
CAMERONI ET AL.
ACE2 binding by Omicron prompted us to question the similarity between the amino acid changes that occur when SARS-CoV-2 is forcibly adapted for growth in mice. Kuiper et al. identified mutations found in a mouse-adapted SARS-CoV-2 isolate, as summarized in Table 1. Four of the changes found in the mouse-adapted virus, Q498R, N501Y, K417N, and Q493R, are also found in the Omicron variant. Position 484 is also mutated, but to a different amino acid: E484A. We also note that many of these mutations are mirrored in sequences pulled from New York City wastewater. Although the origins of the wastewater variants are unknown, one suspected origin is mice.
We have previously speculated the possible origin of Omicron lies in reverse-zoonosis. This is the transmission of the virus from human-to-animal-to-human, with significant mutations added during cross-infection. The similarities between Omicron, the mouse-adapted virus, and the wastewater variants add substance to this hypothesis.

Omicron Convalescent Sera Escape
The next question they address is whether Omicron escapes neutralization by convalescent sera. Omicron strongly evades natural infection. Current data suggests that those who have been previously infected have little to no protection against Omicron infection. To analyze the degree to which the highly mutated Omicron escapes vaccine antibody neutralization, Cameroni et al. created Wuhan S and Omicron S pseudoviruses to test against a wide variety of existing vaccines. They tested the pseudoviruses against the sera from patients vaccinated with Moderna’s mRNA-1273, Pfizer’s BNT162b2, AstraZeneca’s AZD1222, J&J’s Ad26.COV2.S, Russia’s Sputnik-V, and Sinopharm’s BBIBP-CorV.
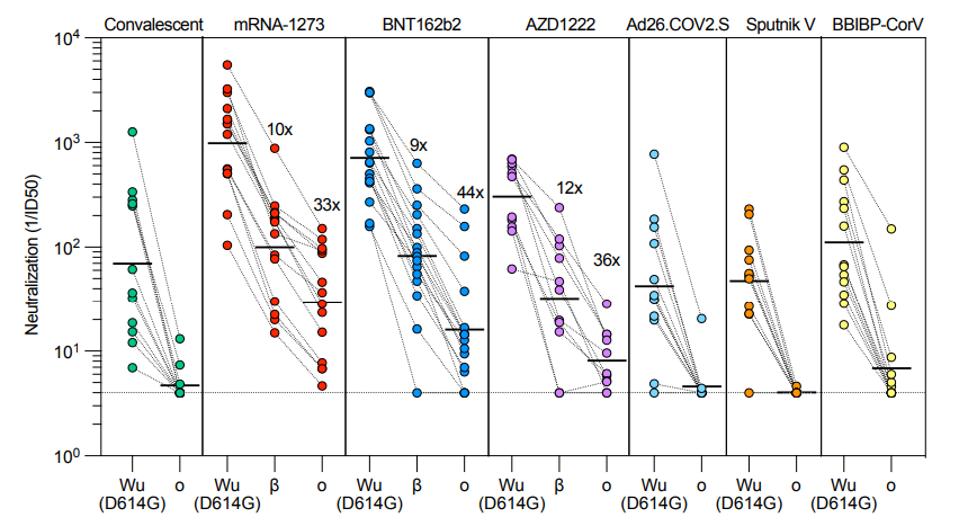
FIGURE 3: Neutralization of Omicron variant pseudovirus by convalescent and vaccinated sera.
CAMERONI ET AL.
In the J&J cohort, no sera samples had neutralizing activity against Omicron. In the Sputnik cohort, only one in the cohort had mild neutralizing activity. And in the Sinopharm cohort, only three had mild neutralizing activity.
The data from figure 4 shows that fully-vaccinated sera with the Moderna vaccine saw a 33-fold drop in neutralization against Omicron, Pfizer a 44-fold drop, and AstraZeneca a 36-fold drop. Notably, Recently released data shows that sera from people who received the third dose lost almost ninety percent of its potency against Omicron after two weeks. After three months, most third-dose sera failed to neutralize at all. This data confirms the epidemiological observations that most fully doubly-vaccinated and triply-vaccinated individuals, including those with prior infection, are susceptible to infection by Omicron within three months of the last boost.
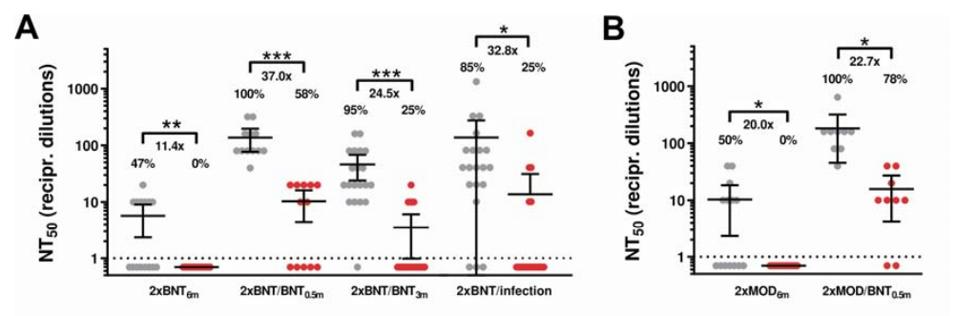
FIGURE 4: Neutralization of SARS-CoV-2 Omicron (red) and Delta (grey) variants by different vaccine regimens.
WILHELM ET AL.
Omicron Monoclonal Antibody Escape
There are currently eight approved or emergency authorized monoclonal antibody treatments in circulation. Most of these and other antibodies in development target the receptor-binding domain, and in particular, the receptor-binding motif from positions 437 to 508. In the Omicron variant, there are ten amino acid substitutions in this region of the Omicron S that are likely to reduce antibody binding and neutralization (Figure 5).
FIGURE 5: ACE2 binding map of different monoclonal antibodies.
CAMERONI ET AL.
Of the eight approved or authorized antibodies, all but sotrovimab completely or almost completely lost their neutralizing activity against the Omicron pseudovirus. A cocktail of cilgavimab and tixagevimab had a reduced potency of 200-fold. Sotrovimab, conversely, shows a drop of 3-fold drop in neutralization potency.
Cameroni et al. also examined a panel of 36 S protein monoclonal antibodies. Four monoclonal antibodies targeted the N-terminal domain, yet none were able to neutralize Omicron. Only three of the 22 antibodies that bind to the receptor-binding motif retained activity including GSK’s S2K146. S2K146 is unique as it makes close contacts with many of the same amino acids as does ACE2 so much so its footprint on the receptor-binding domain closely mimics that of ACE2 itself. Such antibodies may be amongst those preferred for the second generation of anti-SARS-CoV-2 monoclonal antibodies as the virus must bind ACE2 to initiate infection.

FIGURE 6: SARS-CoV-2 Omicron variant N-terminal domain and receptor-binding domain mutations.
CAMERONI ET AL.
We note that Dr. Gideon Schreiber researched the mutated receptor-binding domain to find mutations that would increase the affinity, specifically in ACE2 binding sites. He found mutations that increased the affinity by more than two orders of magnitude. It would seem natural that some of those mutations increase the specificity and affinity of the S2K146 for improved activity and breadth of neutralization.
Continuing with the antibody analysis, of the 9 which targeted other areas of the receptor-binding core, only one retained neutralizing activity. Finally, one antibody, S2H97, was capable of neutralizing Omicron significantly and targets a conserved region noted as site V.

FIGURE 7: (A) Mean Omicron neutralization titers after exposure to monoclonal antibodies and (B) accompanying binding sites.
CAMERONI ET AL.
In all, most monoclonal antibodies were completely ineffective against the Omicron variant. Some, such as sotrovimab, S2K146, and S2X324 retained neutralizing activity, but at a significant reduction. This points to the immune escape capabilities of Omicron via its distinctly mutated S protein and genome.
Conclusion
Many seem surprised at the ability of SARS-CoV-2 to mutate to increase resistance against convalescent sera, most vaccines, and most monoclonal antibodies. However, the surprise was unwarranted for those who realized that coronaviruses have evolved over many millions of years to reinfect hosts over time. Those previously infected with earlier strains of the virus, contrary to our assumptions earlier in the pandemic, are able to be reinfected with Omicron and strains to come.
We previously predicted that SARS-CoV-2 would persist, continue to vary, and evade our natural and adaptive immune responses. We have also learned that SARS-CoV-2 has the potential to become far more lethal than it is today. We reiterate that the sister of this virus, SARS-CoV, and its cousin, MERS-CoV, ranged between 10% and 30% lethality. This is presumably due to slight variations in the structural, nonstructural, and accessory proteins. We must be ever alert now and for many years in the future of the possibility of such changes and their consequences.

