Another Variant Emerges From An Immunocompromised Patient
(Posted on Tuesday, December 21, 2021)
As the pandemic continues, new waves of infections are driven by emerging viral variants with a menagerie of genomic mutations. The exact origins of new mutations are unknown, but one theory is the cultivation of mutations in immunosuppressed Covid-19 patients. In some instances of Covid infection, lingering symptoms may last for months, often in immunocompromised individuals. This allows the virus to effectively adapt within a single patient over time, resulting in potent mutations to the Spike protein and larger genome, particularly in cases when the patient receives any sort of antiviral drug. Here we examine one such case of a 72-year-old patient who was infected continuously for two months, as detailed by Truffot et al.
Patient Background
The patient, a resident of France in his early seventies, had chronic lymphocytic leukemia associated with hypogammaglobinemia for four years prior to his Covid-19 infection. Twenty days prior to the disease course, the patient had received his first dose of the Pfizer/BioNTech vaccine. He additionally received chemotherapy-related monoclonal antibodies, venetoclax and rituximab 17 days prior to disease course.
After experiencing symptoms of coughing, fever, diarrhea, and asthenia, the patient tested positive by PCR test for SARS-CoV-2 infection three days prior to disease course and was given a 700 mg dose of anti-SARS-CoV-2 monoclonal antibody treatment bamlanivimab three days later. For the purposes of this discussion, the administration of bamlanivimab acts as day 0 for the disease course.
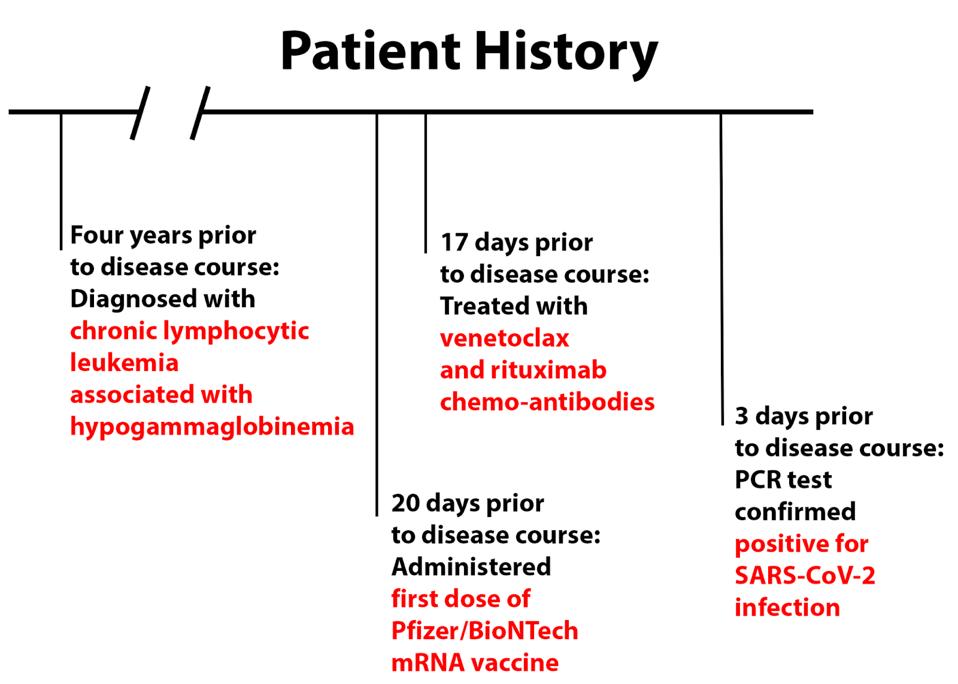
FIGURE 1: Patient history prior to disease course.
ACCESS HEALTH INTERNATIONAL
Patient Treatment
The patient’s condition rapidly deteriorated, requiring hospitalization on day 6 and oxygen assistance and plasma transfusion on day 10. He was transferred to an intensive care unit on day 13 and was treated with high-dose corticosteroids from days 21 to 26. This improved the patient’s baseline condition, but he still required supplemental oxygen, leaving the ICU and returning to the general hospital on day 33, maintaining a high viral load throughout the month.
From days 47-57, the patient received treatment of remdesivir—200 mg on day 47 and 100 mg on the days following. He was discharged to a rehabilitation center following this treatment and tested negative by PCR test on day 61.
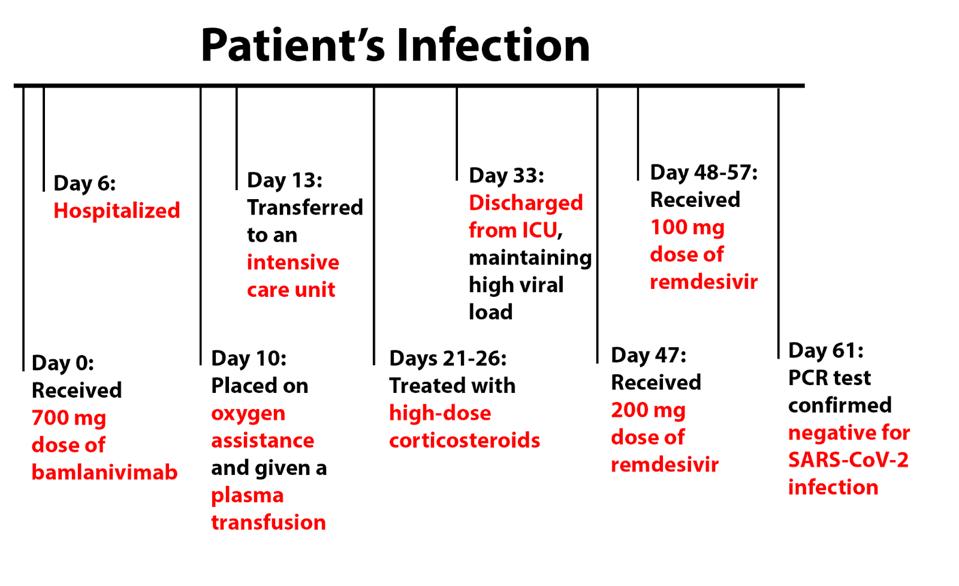
FIGURE 2: Patient viremia and treatment during the disease course.
ACCESS HEALTH INTERNATIONAL
Virus Evolution
Truffot et al. later analyzed 12 samples taken throughout the disease course for genomic mutations by performing whole-genome sequencing. Their analysis revealed the initial infection was a result of the Alpha variant, which drove infections in the winter and spring of 2021. That was the sequence found on day 3. On day 6, sequencing revealed a novel mutation, E484Q, not previously detected in sera samples, which was observed at a frequency of 82%. On day 10, this frequency rose to over 99%. The following day, another novel mutation was detected, Q493R, at a frequency of 64%, which rose to 76% by day 17.
Position 484 in the Spike protein is often mutated in highly immune resistant variants but was never an often observed mutation in the Alpha variant. Additionally, the typical mutation is E484K, whereas the French patient developed E484Q, which is rarer, found mostly in less frequent, regional variants.
Position 493 is interesting as well. Until recently, Q493R in the Spike protein was an uncommon, highly antigenic mutation. Gideon Schreiber described a number of receptor-binding domain mutations for antigenicity, one of which was at positions 493. He described Q493H as a highly antigenic mutation. The Q493R and Q493H mutations are both neutral to positive charges, indicating Q493R should have a similar effect. This means transmissibility for variants that carry this mutation is much higher. The only instance of Q493R in variants of concern is the current Omicron variant which is reigniting Covid-19 infections throughout the world. Its observation in an individual immunocompromised patient is notable, to say the least.
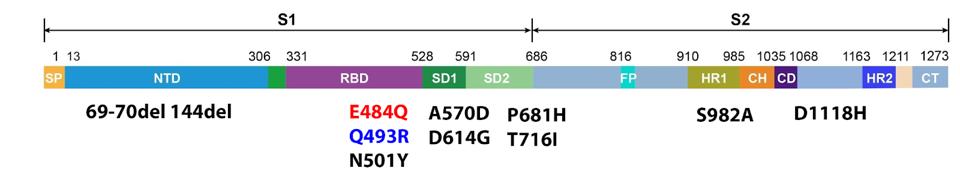
FIGURE 3: Spike mutations sampled from the patient over time. Those in black are those found in the Alpha variant, which the patient was initially infected with. E484Q in red was detected after six days. Q493R in blue was detected after 11 days.
ACCESS HEALTH INTERNATIONAL
Takeaways
This is not the only instance of dangerous mutations developing within an immunocompromised individual. There is a long history of these patients, many of which we have analyzed, including the London, Boston, Pittsburgh, and Italian patients. Recently a 58-year-old patient who underwent a six-month-long infection developed a few questionable mutations in the Spike protein as well, in this instance, E484G and F490L in the receptor-binding domain and several deletions in the N-terminal domain. These mutations all aid in the transmissibility and immune resistance of major variants.
We have recently detailed the potential origins of the new Omicron variant. Understanding how patient zero developed Omicron, or either previous or future variants, may help us gain insight into how to develop strategies to contain these variants before they wreak havoc.
Truffot et al. note that positions 484 and 493 are critical in the bamlanivimab antibody binding process. Giving the immunocompromised patient a steady dose of bamlanivimab may have, in turn, resulted in the development of those mutations.
Genomic surveillance of long-term immunocompromised cases is essential to prevent the development of these mutations. Resources should be devoted to containing infections in these individuals and continuous sequencing on patient sera is vital for genomic surveillance.

