Birth Of The Omicron Family: BA.1, BA.2, BA.3. Each As Different As Alpha Is From Delta.
(Posted on Wednesday, January 26, 2022)
This is the first in a series on the Omicron variant, specifically discussing its derivative lineages: BA.1, BA.2, and BA.3. We have previously speculated on the potential origin of Omicron in an earlier article.
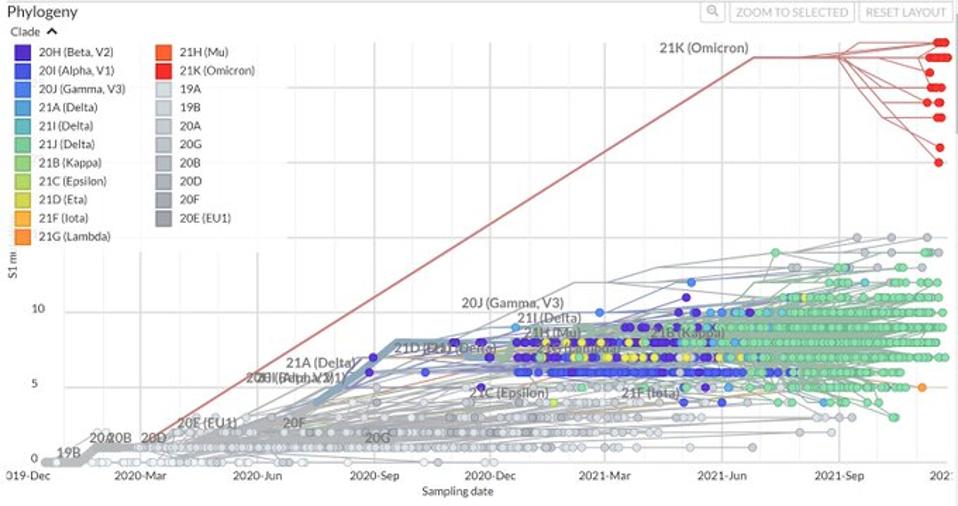
Time course for the evolution of significant SARS-CoV-2 variants, note the considerable divergence of the Omicron variants from all other variants, branching off from other variants as early as March 2020.
NEXTSTRAIN.ORG
The Omicron variant has continued to surprise by its sudden appearance, its evident ability to evade both vaccines and immune responses, and its rapid spread worldwide. There’s another surprise buried in the emergence of Omicron. The variant is not a single strain, but rather a family of three: BA.1, BA.2, and BA.3.
We should take all three sublineages seriously. BA.1 is the most prolific sublineage, detected in most countries worldwide and currently accounting for 99% of cases in the United States. BA.2 is less prolific, but has overtaken BA.1 in Denmark, Nepal, and the Philippines as the most frequently detected variant, and has a minor presence in India, the United Kingdom, and several other countries. The third, BA.3, is yet to take off globally, only accounting for several hundred cases at the most.
Each of these variants is as different from one another as Alpha, Beta, Gamma, and Delta are from one another. What this means for the current state of the pandemic is uncertain. However, one thing is clear: SARS-CoV-2 has an enormous capacity not only to continue to produce new variants, but variants that surprise us both in their number and their biological properties.
Here, we introduce the Omicron family of variants. More detailed descriptions of each strain will follow this introduction. We have already described BA.1 in a previous story under the moniker of “Omicron” before the family was apparent. In this first entry, we will introduce the three, how they diverged from the Wuhan strain, and how they compare to each other.
As with any variant of SARS-CoV-2, we return to the source of the virus: the wild-type Wuhan strain. This is the blueprint for all variants to come. The significant variant has no Greek letter name, but rapidly displaced the Wuhan almost everywhere in the world except East Africa. Here we call it the Triad, a name that denotes three mutations, the D614G mutation in the Spike protein, the P323L mutation in the NSP12 polymerase, and the C241U noncoding mutation in the 5’ end. The Triad is the founding variant of all variants of concern viruses.
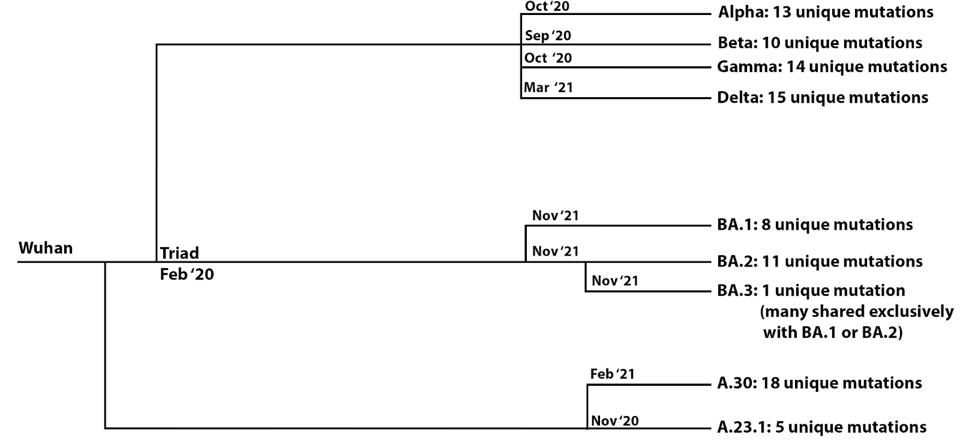
FIGURE 1: Evolutionary distances between major variants of concern, the Omicron family of viruses, and select African “A” viruses. The number of unique mutations is noted for each variant, illustrating how different the sublineages are from each
ACCESS HEALTH INTERNATIONALWe note that the Omicron variant significantly differs from other major variants of concern. We identify a precursor, which we call the Omicron parent, likely to have risen about March of 2021. The three sublineages share 39 mutations, which we include as the presumptive “Omicron parent.” The Omicron parent diverged into the Omicron family: BA.1, which contains an additional 20 mutations; BA. 2, which includes an additional 27; and BA.3, which consists of an additional 13. Remarkably, all the family members were detected simultaneously in South Africa, although they likely diverged from one several months previously. This is a unique example of such highly divergent strains appearing in a population simultaneously.
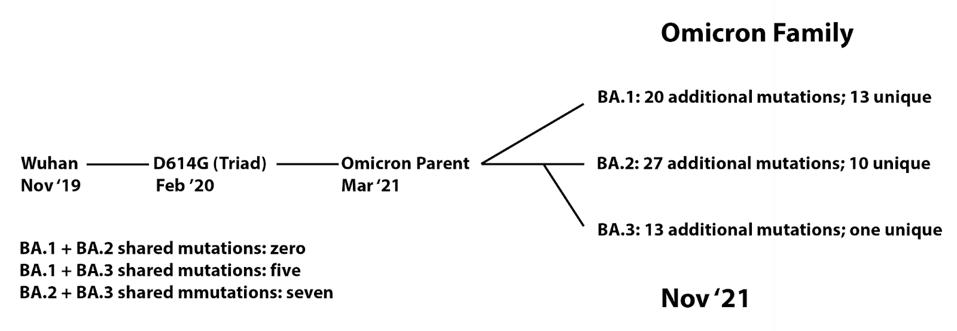
FIGURE 2: Lineage of Omicron family viruses. The date included is when that specific variant was first collected.
ACCESS HEALTH INTERNATIONALOf the 39 mutations introduced in the Omicron precursor, most are located in the Spike protein. This is the virus region that makes contact with the host cell ACE2 receptor and plays a significant role in transmissibility and evasion of neutralizing antibodies. Non-S-protein mutations are sprinkled throughout and may play an equally important role in the transmission and pathogenesis of the virus. Figure 3 displays the similarities and differences amidst the three sublineages.
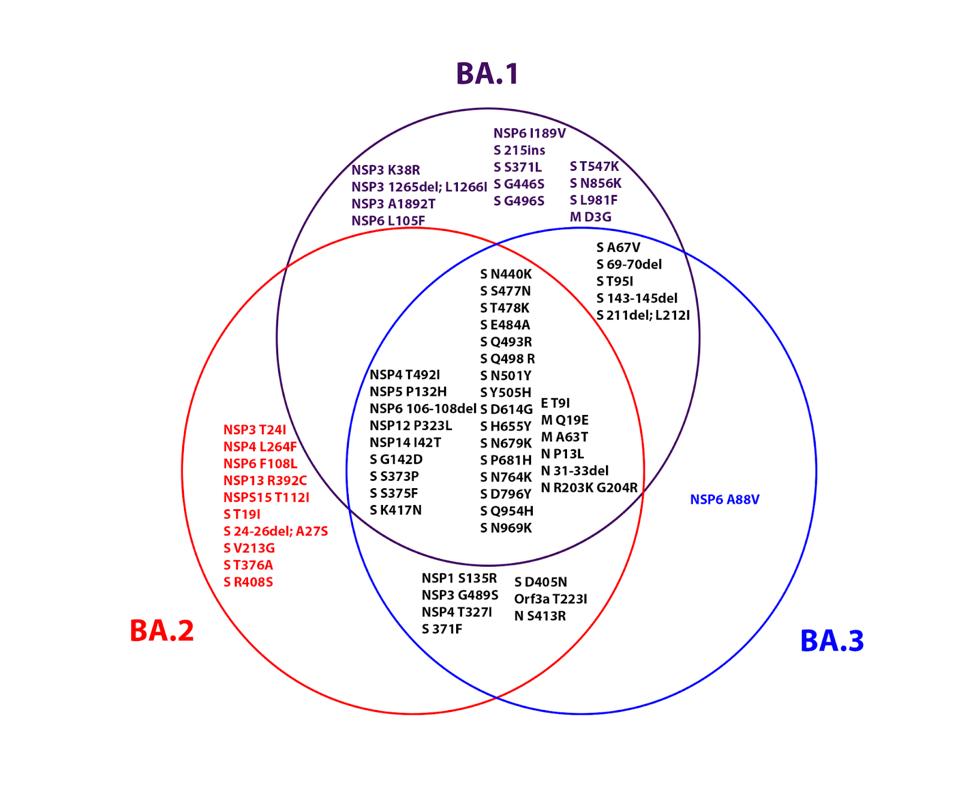
FIGURE 3: Venn diagram showing the similarities and differences between the three Omicron family viruses.
ACCESS HEALTH INTERNATIONALThe largest single class of mutations is shared amongst all three Omicron family members. Figure 3 emphasizes the differences amongst the three variants as well. We reiterate that BA.1, BA.2, and BA.3 differ from each other as much as the Alpha, Beta, Gamma, and Delta variants differ from one another. Several mutations differ between the three as well. We suspect that these differences will be reflected in the septic characteristics of each variant affecting growth rate, suppression of innate immunity, virulence, and vaccine evasion.
Omicron (BA.1)is already known to have some unusual biological properties relative to the other variants. Most strikingly, it is 2.7-3.7-fold more transmissible than Delta. The origin of the increased transmissibility is somewhat of a mystery, as the concentration of virus in nasal sections is not exceptionally high, and is, in fact, lower than that of Delta. Moreover, the affinity of the BA.1 Spike for the ACE2 receptor is only twice as high as the Wuhan strain. We speculate that the increased transmissibility may be in the mutations found in the nonstructural, structural, and accessory proteins found throughout the genome.
Another biological property is how Omicron enters the cell. The Wuhan wildtype and all previous variants enter primary via membrane-to-membrane fusion. Omicron seems to prefer an endosomal route similar to SARS-CoV (the SARS virus) but different from all previously characterized variants. Omicron does not induce cell to cell fusion (syncytial formation) in cell culture, consistent with endosomal entry.
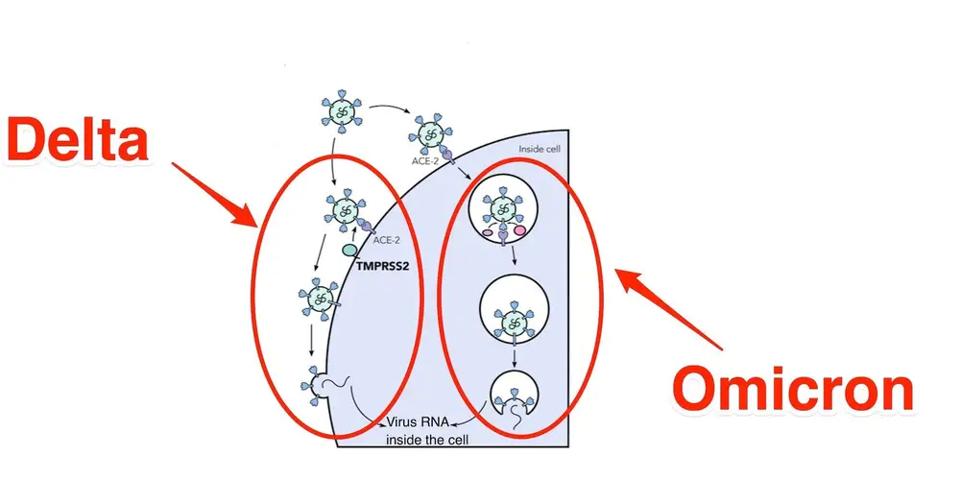
FIGURE 4: Membrane to membrane vs. endosomal entry of SARS-CoV-2 in Delta vs. Omicron
GEORGINA BROWN/JOE GROVE/INSIDERThe fundamental change in cell entry may be explained by a set of mutations described by Martin et al. They describe three clusters of mutations that appear in SARS-CoV-2 variant very rarely, if at all, based on the 7.4 million SARS-CoV-2 sequences in GISAID.
The first cluster is composed of receptor-binding mutations G339D, S371F/L, S373P, and S375F. The second cluster is composed of receptor-binding domain mutations Q493R, G496S, Q498R, and Y505H. The third is composed of fusion domain mutations N764K, N856K, Q954H, N969K, and L981F. The combination of these clusters may well lead to the modified preference of cell entry. We note that three of these mutations, G496S, N856K, and L981F, are found only in BA.1, not BA.2 or BA.3. This may impact the viral fitness of BA.1 relative to its sibling sublineages. How and to what extent they are affected, we do not yet know.
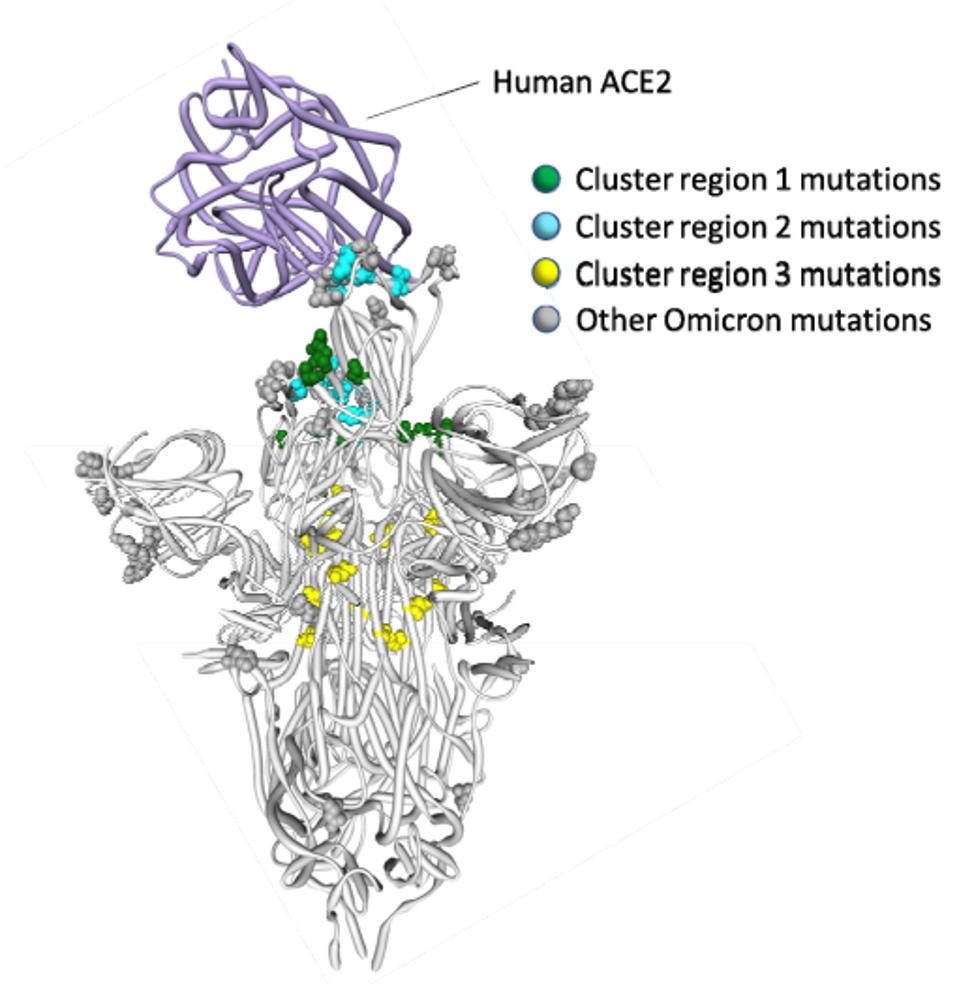
FIGURE 5: Three Spike cluster mutations in Omicron as described by Martin et al.
MARTIN ET AL.This series aims to understand Omicron and its sublineages in more detail, as they are by far the most transmissible version of SARS-CoV-2 to date, but fortunately less pathogenic—at least for well vaccinated healthy young adults. We will examine the mutations in BA.2 and BA.3 in stories to follow soon.

