Rabbitpox: the Story of a Specialized Killer
(Posted on Monday, June 6, 2022)
With poxviruses now back on the radar, we must consider the outstanding threat of possible human pathogens in much greater detail. It is critically important to understand not only how poxviruses jump from one species to another but also how they evolve across generations. As we reported in part one of this series, the “arms race” between the myxoma virus and its primary hosts, rabbits, has been one of the most well studied examples of how viruses mutate and coevolve with their environment. Scientists initially introduced the myxoma virus to European rabbits to control their growing populations in parts of Europe and Australia. This virus that was naturally found in South American sylvilagus rabbits killed 99% of the European rabbits it infected. Over time, surviving generations developed mechanisms to resist viral infection, which reduced the virus’ fatality rate by nearly 50%.
Just as European rabbit populations were evolving, however, so was the virus. Newer strains of the myxoma virus, in fact, increasingly suppress the immune systems of their rabbit hosts. Like any other virus, the myxoma virus must overcome the host’s immunity barriers to replicate within cells successfully. As the cell’s first line of defense, innate immune responses block viral replication by inducing the expression of interferon-stimulated genes. Suppressing these innate immune responses, as well as later immune mechanisms, determines how well the virus can replicate and infect others.
Expressing host range factors is one way that viruses can evade immune detection. The function and prevalence of these virus-encoded proteins varies across different cell types, hosts and species. For example, poxviruses are the only viruses that express host range factors from the C7L family. The C7 sequence was originally identified in the vaccinia virus that causes smallpox. Deleting the C7L gene from the smallpox virus resulted in a mutated virus that could no longer replicate in human and other animal cells. Without the C7L protein, experimentally enhancing the expression of interferon-stimulated genes could be a potent treatment against smallpox, but reintroducing C7L blunted this effect. This finding revealed that C7L and other similar host range factors may interfere the effectiveness of interferon-stimulated genes recruited during innate immunity.
C7L and its homologs are expressed in nearly every poxvirus that infects mammals, including the myxoma rabbitpox virus. The C7L family consists of all the protein homologs across different poxviruses that exhibit a similar genetic sequence to the original smallpox C7L protein. Despite the similarity of their genetic sequences, this family of host range factors can be divided into two major groups based on their role in viral replication. The first group, which includes most members of this family, determines how well the virus can replicate and therefore are not functionally different from the C7L smallpox protein. The second group does not play a critical role in replication in most cells. Within this group, however, a member of the C7L family may enhance replication in one species but not others.
The Crossover
In fall 2018, hundreds of hares from the Iberian Peninsula seemingly died overnight from a mysterious virus. A genetic analysis found that this novel virus, which researchers called MYXV-Tol, was very similar to the original South American rabbitpox virus, suggesting that this virus was a novel strain of the myxoma virus. Given that these hares were previously resistant to rabbitpox, what changed? The key difference was that the new MYXV-Tol virus acquired a unique “gene cassette” through DNA recombination. Gaining approximately 2,800 base pairs from some unknown poxvirus, this hybrid strain expresses several new genes compared to the original virus. The placement of these new protein-encoding genes is shown below in Figure 1.
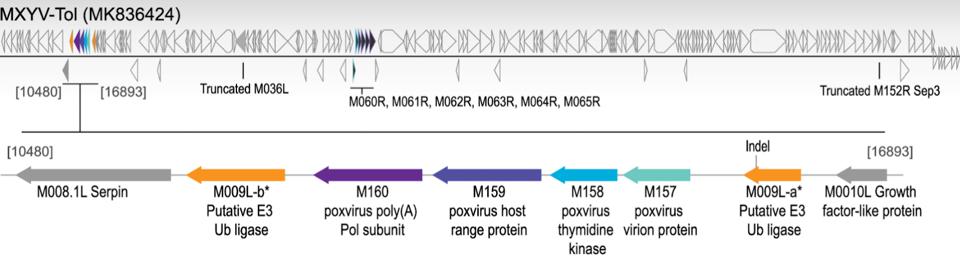
Figure 1: A representation of MXYV-Tol viral genome. New genes that were acquired from DNA FROM: “IDENTIFICATION OF A NOVEL MYXOMA VIRUS C7-LIKE HOST RANGE FACTOR THAT ENABLED A SPECIES LEAP FROM RABBITS TO HARES” ÁGUEDA-PINTO ET AL. 2022
Now, in a recent report, Águeda-Pinto et al. from Arizona State University singled out a key gene from the novel recombinant virus, called M159, that may have enabled the myxoma virus to jump from rabbits to hares. Characterized as a host range factor, understanding the role of this protein may provide insight into how poxviruses bypass innate immunity to successfully infect hosts.
To investigate the role of this protein, Águeda-Pinto et al. knocked out, or removed, the M159 sequence from the myxoma virus they detected in the hares. How well this knockout virus could replicate within the extracted hare cells was then compared to unchanged versions of the virus.
Subsequent findings from this experiment confirmed that the replication of the myxoma-like virus within hare cells depended on the expression of M159 proteins. Removing this gene prevented the virus from infecting or replicating within host hare cells. Exposure to the unchanged virus, however, did lead to infection. When researchers inserted the M159 protein sequence into an alternate South American strain of the myxoma virus, the virus increasingly replicated within the cell samples, even though Iberian hares are not normally susceptible to this viral strain. Águeda-Pinto et al. reported that the specific backbone of the virus, in addition to the presence of M159, is critical for infecting these hare cells.
Investigators also found that the addition of M159 enabled the virus to infect human cancer cells. Humans are not normally susceptible to rabbitpox. However, when Águeda-Pinto et al. exposed pancreatic and melanoma cancer cells to M159-enhanced strains of the myxoma virus, they were able to engineer a tumor-fighting virus that successfully replicated within cells. The ability for M159 to enhance viral replication may provide a useful target for fighting cancers in humans.
How is M159 enhancing viral replication? In vaccinia viruses, these proteins have been shown to suppress early innate immune responses to promote viral replication. How this happens has yet to be well understood. What we do know is that without this immunosuppressive gene, a cell can more easily shut down the invading virus and block further replication.
Investigators speculate that M159 likely binds to and interferes with interferon-stimulated gene products that are recruited for innate immune responses to viral pathogens. Like other members of the C7 family, M159 folds into a protein scaffold that can bind to specific host-generated antiviral proteins. The structure of this protein looks like a hand folded upwards.
Despite being a member of the C7 host range factor family, M159’s genetic sequence only slightly resembles that of C7L. In fact, this protein only shares five amino acids with C7L. Like a hand with many fingers, only the fingertips are conserved. The rest of the hand has an entirely different protein sequence. However, it is the conserved fingertips of this protein that are reportedly critical for binding host-generated antiviral proteins.
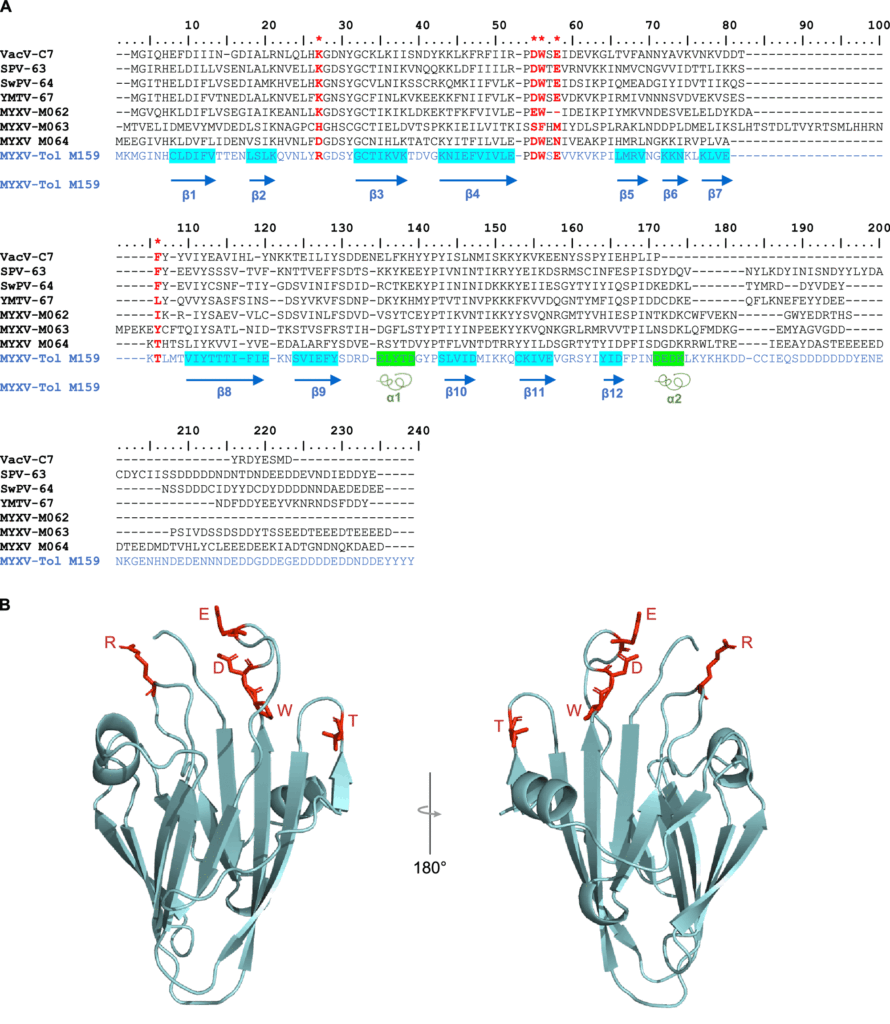
Figure 2: An illustration of the M159 protein gene identified in the MYXV-Tol rabbitpox variant that FROM: “IDENTIFICATION OF A NOVEL MYXOMA VIRUS C7-LIKE HOST RANGE FACTOR THAT ENABLED A SPECIES LEAP FROM RABBITS TO HARES” ÁGUEDA-PINTO ET AL. 2022
Unlike other DNA viruses that need to enter the nucleus to replicate, poxviruses can operate directly from the cell’s cytoplasm. At early, intermediate and late stages of infection, these viruses capitalize on different gene regulatory mechanisms to enhance replication. Once the virus binds to and invades a host cell, a viral inner core is released into the cytoplasm. This prompts the transcription of early viral mRNA proteins, including host range factors that can evade the immune system and facilitate the synthesis of viral RNA products. Intermediate and late mRNAs, then, synthesize products needed to assemble new versions of the virus.
To determine which stage M159 is implicated, Águeda-Pinto et al. examined the expression of known early and late viral proteins in hare cells following exposure to both changed and uncharged versions of the myxoma-like virus. Gene expression of early viral proteins was unaltered with exposure to various forms of this virus. Deletion of M159, however, prevented the formation of new viruses, suggesting that this viral protein is involved somewhere between early and mid-to-late stages of infection.
Yet, there is still another pressing question: did the addition of M159 make European rabbits more vulnerable to severe infection? To their surprise, Águeda-Pinto et al. found that the presence of M159 did not enhance viral infection and replication in rabbit cells. Although it has been reported that European rabbits can contract the myxoma-like virus, the rate that they are infected is much less frequent than Iberian hares. M159 is likely a host range factor that is specific to hare species.
We should not rush to assume that the addition of M159 in the novel myxoma strain was the only change that enabled the virus to cross the species barrier, however. This recombinant virus expresses several new genes, in addition to M159, that may have enhanced the virulence of rabbitpox in hares. Since the full nucleotide sequence of the myxoma-like virus has yet to be elucidated, there may have been several factors that contributed to the enhanced infectability of the Iberian hares.
As Grant McFadden, director of the Biodesign Center for Immunotherapy, Vaccines and Virotherapy at ASU and coauthor of this study, says, “Every time a virus leaps from one host species into another, we learn something new about Mother Nature.” Sometimes the things we learn, however, only lead to more questions. What does this new myxoma strain mean for other animal species that live alongside these hares and rabbits? Can we predict how the virus will evolve in the future to prevent other species from being infected? These and many other questions exemplify why it is important to identify viral spillover events as they occur. The next pandemic may likely be a zoonotic poxvirus.

