How Hepatitis D Virus Hijacks Host Replication Machinery
(Posted on Wednesday, June 14, 2023)
Originally published on Forbes on June 9, 2023.
This story is part of a larger series on viroids and virusoids, small infectious RNAs. It is also the fifth installment in a series on hepatitis D virus, a virusoid-like pathogen that causes serious human disease. You may read the others on Forbes or www.williamhaseltine.com.
All viruses encode an enzyme capable of replicating their genome, be it DNA or RNA. Without such an enzyme, the virus is rendered impotent — even if it manages to infiltrate a host cell, it cannot replicate. Viroids and virusoids, including hepatitis D, are the exception. These small, circular RNA pathogens do not encode replication enzymes of their own. Despite this, they manage to replicate without issue. How? The simple answer is that they hijack the replication machinery of the host cell. Although most viroids and virusoids do not encode proteins, hepatitis D virus does. The hepatitis D protein —known as hepatitis D virus antigen (HDAg)— modifies the relevant cellular machinery to its own advantage. In particular, it interacts with host DNA-dependent RNA polymerases (Pol I, Pol II, and Pol III). What follows is an overview of this strategy and its importance to the hepatitis D life cycle (Figure 1). Understanding these interactions opens the opportunity for new hepatitis-D-specific antiviral drugs.
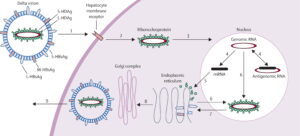
FIGURE 1. Schematic representation of the hepatitis Delta virus (HDV) lifecycle. SOURCE: Hughes et al. 2011 https://doi.org/10.1016/S0140-6736(10)61931-9
What Are Host RNA Polymerases?
DNA-dependent RNA polymerases are enzymes that a cell uses to copy the information in DNA to RNA. There are three human RNA polymerases, designated RNA polymerase I, II, and III.
RNA Polymerase I
RNA polymerase I is a large protein composed of 14 different subunits, known as polypeptides. It contributes to cell division by creating many of the components of ribosomes, the protein-producing machinery of cells. Think of ribosomes as translators; as they traverse a strand of RNA, they convert the nucleotides that make up the RNA into amino acids. RNA in, proteins out.
RNA polymerase I produces these ribosomal pieces —ribosomal ribonucleic acids (rRNAs), to use the technical term—by transcribing a special kind of DNA called ribosomal DNA (rDNA). The ribosomal RNA transcribed by RNA polymerase I joins up with ribosomal proteins to form a complete ribosome.
Transcription of ribosomal DNA into ribosomal RNA by RNA polymerase I happens in a separate subcompartment of the nucleus known as the nucleolus.
RNA Polymerase II
RNA polymerase II is also a multi-protein complex, made up of 12 different subunits. Five of its subunits are shared with RNA polymerase I and III. Despite this, its function is very different; where RNA polymerase I exclusively transcribes ribosomal DNA, RNA polymerase II transcribes the protein-coding sections of ordinary DNA into messenger RNA. These messenger RNAs are then used as blueprints for the synthesis of proteins.
Transcription of DNA by RNA polymerase II occurs in the nucleoplasm.
RNA Polymerase III
RNA polymerase III is the largest of the three RNA polymerases, composed of 17 subunits. Again, it shares five of its subunits with the other two RNA polymerases. And like the other two polymerases, it also serves to facilitate transcription of DNA into particular kinds of RNA. Specifically, it transcribes DNA to produce 5S ribosomal RNA —the only ribosomal RNA molecule not transcribed by RNA polymerase I— and to produce transfer RNA (tRNA).
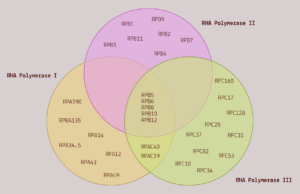
FIGURE 2. A Venn diagram comparing the subunit composition of the three RNA polymerases. SOURCE: ACCESS Health International
In addition to the aforementioned RNA polymerases, there are two more (Pol IV and Pol V). However, these are exclusively found in plants. Since hepatitis D is an animal pathogen, the two plant RNA polymerases are not relevant.
Hepatitis D-RNA Polymerase Interactions
Recall that DNA-dependent RNA polymerases usually take DNA as a template and convert it into one or another form of RNA. Hepatitis D virus, however, is made up exclusively of RNA. This means that the pathogen somehow appropriates the polymerases into using RNA as a template — RNA to RNA, instead of DNA to RNA. How does it achieve this act of subversion?
Macnaughton and Lai propose two possible explanations, which, it should be noted, are not mutually exclusive. The first is that the small protein of hepatitis D virus binds directly to the RNA polymerases, initiating replication and transcription. The other possibility is that the quasi-double stranded secondary structure of hepatitis D virus —a result of 70% sequence self-complementarity— is mistaken for DNA, thereby hijacking RNA polymerases and transcription factors that usually would not bind to RNA.
Small Hepatitis D Protein
Evidence for the first explanation is abundant. For one, the small hepatitis D protein interacts with nine of the 12 RNA polymerase II subunits. Along with these nine subunits, the small protein was also found to interact with 65 other host proteins involved in transcription. Many of these are also known to bind RNA polymerase II. It is possible that, after binding to RNA polymerase II, these proteins then also link to the small protein to further replication.
Another point in favor of this possibility is that the small protein of hepatitis D binds to a section of RNA polymerase II known as the “clamp”, which acts as a claw that allows the polymerase to latch onto DNA (Figure 3). In binding to this region, the small protein loosens the grip of the clamp. The effect of this is twofold. On the one hand, loosening the clamp speeds up the rate of transcription, and by extension, the rate of replication. On the other hand, it decreases fidelity of transcription. This is not necessarily a bad thing. On the contrary, it may be the mechanism by which the small protein is able to hijack RNA polymerase II: decreased fidelity may represent a relaxation of the usual template requirements. Indeed, Yamaguchi et al. note, “By reducing transcriptional fidelity in terms of not only discrimination of incoming nucleotides but also recognition of templates, HDAg may facilitate the unusual RNA-dependent RNA synthesis by Pol II.”
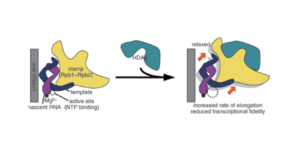
FIGURE 3. Schematic representation of hepatitis D antigen (HDAg) as it attaches to RNA polymerase II. SOURCE: Yamaguchi et al. 2007 https://doi.org/10.1111/j.1365-2443.2007.01094.x
Finally, the small hepatitis D protein shares some of its sequence with a region of a protein complex called negative elongation factor (NELF). Negative elongation factor binds to RNA polymerase II and represses transcription. In order for transcription to begin, negative elongation needs to first be removed — think of the complex as a kind of gatekeeper, preventing RNA polymerase II from activating in instances it shouldn’t. As the small protein binds to RNA polymerase II, it displaces the negative elongation factor, kick-starting the polymerase into transcription mode.
The next article in this series will look at the possibility that the hepatitis D virus genome itself interacts directly with the RNA polymerases, independently of the small antigen protein.

