Molecular Acrobatics: A Look At Hepatitis D Virus Replication
(Posted on Tuesday, June 20, 2023)
Originally published on Forbes on 6/12/2023
This story is part of a larger series on viroids and virusoids, small infectious RNAs. It is also the sixth installment in a series on hepatitis D virus, a virusoid-like pathogen that causes serious human disease. You may read the others on Forbes or www.williamhaseltine.com.
Viroids and virusoids are small, circular RNA pathogens. They have a very unique ability: with only a tiny amount of genetic information, they are able to transmit between hosts, infect cells, and cause disease. Viruses and other microbes also manage to do this, but by comparison they are veritable giants. The virus that causes Covid-19, for example, depends on roughly 30,000 nucleotides —the building blocks of ribonucleic acid (RNA)— to achieve its dirty work. The average viroid or virusoid, in contrast, weighs in at only 200 to 450 nucleotides.
As well as being larger, most viruses encode multiple proteins to help them replicate and, in especially nefarious cases, evade and suppress the host immune system — they come prepared, toolbox in hand. The vast majority of viroids and virusoids do not encode any proteins of their own. Instead, they depend entirely on the host to help them replicate. Where viruses bring their own tools, viroids and virusoids simply appropriate the tools already present.
Some of the most important cellular tools for viroid and virusoid replication are host enzymes called DNA-dependent RNA polymerases. These are proteins that usually copy sections of DNA into RNA; DNA in, RNA out. Somehow, viroids and virusoids manage to hijack these polymerases, fooling the proteins into transcribing viroid RNA genomes instead of their usual DNA targets; RNA in, RNA out. This act of molecular acrobatics allows the pathogens to create multiple new copies of their genome, each of which can then go on to become a fully infectious particle.
Hepatitis D “virus”, despite the name, is a virusoid. Like most other virusoids, it depends on host RNA polymerases for replication. Unlike most other virusoids, it encodes proteins of its own. These proteins contribute to the replication process. In a previous article, we reviewed how the small hepatitis antigen protein (S-HDAg) binds to and alters RNA polymerase II, kickstarting replication. Here, we look at an alternative possibility: perhaps sections of the hepatitis D genome alone, independently of the small hepatitis D protein, suffice to pirate the host RNA polymerases. This would line up with what we know about other viroids and virusoids, which manage to replicate without encoding any proteins.
No Proteins Needed? Hepatitis D Stem-Loop Regions
For a long time it was suspected that RNA polymerase II —one of three DNA-dependent RNA polymerases present in humans— was associated with the replication of hepatitis D virus. Anytime alpha (α) amanitin, a toxin that interrupts RNA polymerase II, was introduced, replication of hepatitis D virus was also inhibited. Exactly how and where polymerase II recognizes the hepatitis D genome, however, was unclear.
As with many viroids and virusoids, the hepatitis D virus genome has a high degree of self-complementarity — around 70% of the nucleotides pair up. Since the genome is circular, the pairing of nucleotides causes the circle to fold in on itself, resulting in a rod-like structure (Figure 1). In areas of low or no sequence self-complementarity, single bulges or loops remain. This includes stem loop structures at each end.
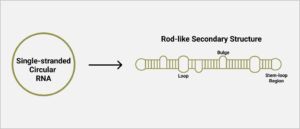
FIGURE 1. Schematic representation of hepatitis D virus secondary structure. SOURCE: ACCESS Health International
To find out which parts of the hepatitis D virus genome interact with RNA polymerase II, Greco-Stewart et al. broke the genome down into three domains: a 213-nucleotide-long section corresponding to the left stem-loop region (nucleotides 687 to 900); a section corresponding to the quasi-double-stranded middle section (nucleotides 681 to 54 and nucleotides 900 to 1540); and a 199-nucleotide-long section corresponding to the right stem-loop region (nucleotides 1540 to 60). This was done for both the positive-strand RNA as well as the negative-strand RNA.
Exposing these three regions to RNA polymerase II yielded a surprising result: only the stem-loop regions at the ends of the hepatitis D genome interacted with the host enzyme (Figure 2B). This was the case for both polarities of the genome. The middle section, on the other hand, showed no indication of interacting with the host enzyme. This suggests that the RNA polymerase II interaction sites are between nucleotides 1540 to 60 and nucleotides 687 to 900 of the hepatitis D virus RNA. These observations also suggest that the cellular DNA-dependent RNA-polymerase recognizes the specific structure, not the nucleotide sequences themselves as the sequences differ not only between the two ends but also between the stem loop corresponding to the positive and negative versions of the genomic RNA.
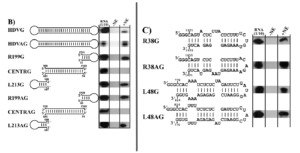
FIGURE 2. Interaction of RNA polymerase II with HDV-derived RNAs. (B) Interaction of RNA polymerase II with: hepatitis D virus genome (HDVG); hepatitis D virus antigenome (HDVAG); right stem-loop section, 199 nucleotides, genomic strand (R199G); central section, genomic strand (CENTRG); left stem-loop section 213 nucleotides, genomic strand (L213G); right stem loop, 199 nucleotides, antigenomic strand (R199AG); central section, antigenomic strand (CENTRAG); left stem-loop section 213 nucleotides, antigenomic strand (L213AG). (C) Interaction of RNAP II with the extremities of the rod-like HDV secondary structure of both polarities. SOURCE: Greco-Stewart et al. 2007 https://doi.org/10.1016/j.virol.2006.08.010
To hone in on the exact location of the interaction sites, the researchers broke down the stem-loop regions into smaller sections, testing each for their ability to interact with RNA polymerase II. They found that a 38-nucleotide sequence on the tip of the right stem-loop region and a 48-nucleotide sequence on the tip of the left stem-loop region were particularly integral to successful binding (Figure 2C). Indeed certain nucleotide sequences within these regions proved to be fully conserved across 81 different variants of the hepatitis D virus and there seems to be high selective pressure to keep the rod-like secondary structure of these regions.
Along with RNA polymerase II, RNA polymerases I and III are also implicated in hepatitis D replication. Building on their previous work, Greco-Stewart et al. confirmed that hepatitis D virus interacts with RNA polymerases I and III; it associates with transcription initiation factor TFIID subunit 1 (TAF1) and with the DNA-directed RNA polymerase III subunit RPC10 (POLR3K). Transcription initiation factor TFIID subunit 1 is a subunit of a protein complex called selective factor 1 (SL1). Selective factor 1 binds to the target DNA and attracts another protein to which RNA polymerase I can bind, initiating transcription. POLR3K —also known as RPC10— is a subunit of RNA polymerase III that copies cellular DNA into transfer RNAs. It also plays an important role in termination and initiation of transcription.
As with RNA polymerase II, polymerases I and III associate with both polarities of hepatitis D RNA in liver cell lines. And the two polymerases also interact exclusively with the stem-loop regions of the hepatitis D genome (Figure 3).
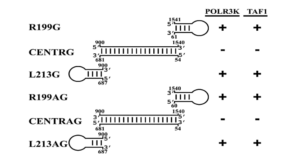
FIGURE 3. Compilation of the interaction of various HDV-derived RNAs with both TAF1 and POLR3K. Stem-loop domains: R199G, R199AG, L213G, and L213AG. Central domains: CENTRG and CENTRAG. G= genomic, AG = antigenomic. SOURCE: Greco-Stewart et al. 2009 https://doi.org/10.1016/j.virol.2009.02.007
Connecting the Dots: TATA-Binding Protein?
All three human RNA polymerases interact with the stem-loop regions of the hepatitis D genome; might there be some common feature shared by the polymerases that enables this interaction?
The answer may lie in the TATA-binding protein (TBP). This protein is common to all three of the RNA polymerases and plays a central role in initiating transcription. Indeed, binding of the TATA-binding protein to the target DNA is the first step in transcription; it provides an anchor site for additional “transcription factors” —other proteins involved in transcription— and it recruits these proteins into action.
Greco-Stewart et al. mention, “[TATA-binding protein] can recognize both TATA and TATA-less promoters dependent on accessory transcription factors, and its affinity for DNA templates is mediated by electrostatic interactions with the phosphate backbone, which could account for its ability to bind HDV RNA despite the absence of known DNA promoter features.”
Implications
Clearly, viroids and virusoids punch well above their weight. With a few hundred nucleotides, they achieve what other viruses need tens or hundreds of thousands of nucleotides to do. Understanding these fascinating entities, and how they achieve so much with so little, may help us design more efficient drugs and inform novel messenger RNA therapies.

