Hepatitis D Virus-Like Agents Are Ubiquitous Throughout The Animal Kingdom
(Posted on Tuesday, June 27, 2023)
This article was originally published on Forbes on June 22, 2023.
This story is part of a larger series on viroids and virusoids, small infectious RNAs. It is also the eighth installment in a series on hepatitis D virus, a virusoid-like pathogen that causes serious human disease. You may read the others on Forbes or www.
Hepatitis D virus has generally been considered an exclusively human pathogen, but a fast-growing body of research suggests otherwise. Hepatitis D virus-like agents seem to be active in a range of different animal species, from fish all the way to termites. Such findings call for a reevaluation of previous assumptions and a recognition that hepatitis D-like viruses may be significantly more pervasive than initially thought.
Hepatitis D “virus” is a tiny, infectious RNA pathogen. Far smaller than any known virus, it more closely resembles subviral agents known as viroids and virusoids. These are characterized by their circular genomes and lack of replication proteins: where viruses encode proteins to help them replicate their genetic information, viroids and virusoids instead hijack host-cell proteins. The same is true of hepatitis D virus. And like other virusoids, it is also understood to be dependent on the envelope proteins of a helper virus —in this case hepatitis B virus— for transmission between hosts and between cells.
Metatranscriptomics: Fishing for RNA Sequences
Central to these new findings is a technique called metatranscriptomics. Pioneered
Metatranscriptomics allows researchers to extract massive amounts of RNA from environmental samples that would otherwise be difficult to study; in a sense, it acts as a giant sieve that filters out everything that isn’t RNA content.
Duck, Duck, Hepatitis D?
One of the first traces of hepatitis D virus-like agents in non-human hosts was discovered in 2018 by a group of researchers based in Australia. As part of a broader avian metatranscriptome study, Wille et al. analyzed throat and anal swabs collected from ducks caught at a water-treatment plant in Melbourne. They noticed that some of the RNA resembled that of human hepatitis D virus.
The nucleotide contents didn’t quite match up — human hepatitis D virus has a guanine (G) and cytosine (C) content of 60%, whereas the guanine and cytosine content of the new sequences was only around 51%. This aside, the two RNA sequences shared many features suggestive of common ancestry. For one, the avian hepatitis D virus-like RNA was circular and folded in on itself to form an unbranched rod-like secondary structure, a key characteristic of human hepatitis D virus. The hepatitis D virus-like genome discovered in the duck samples also had an open reading frame with the capacity to express an 185-amino-acid-long protein. The location of this open reading frame within the genome very closely mirrors the location of the open reading frame seen in human hepatitis D virus (Figure 1). On top of this, certain sequences of the avian hepatitis D virus-like agents resemble those corresponding to the genomic and antigenomic ribozymes found in human hepatitis D virus. Indeed, both of these potential avian ribozymes had secondary structures very similar to those of human hepatitis D virus ribozymes (Figure 2).
Curiously, none of the duck samples showed any signs of infection with duck hepatitis B virus. One possible explanation for this is that non-human hepatitis D virus-like agents may take advantage of other viruses to make infectious particles and to transmit from cell to cell. This explanation is supported by later work, which showed that, even in humans, hepatitis B virus is not the only one that can function as a helper virus.
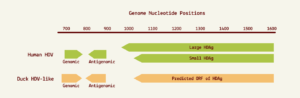
FIGURE 1. Location of genomic and antigenomic ribozyme sequences, and the predicted ORF of the delta antigen in the avian HDV-like genome compared to their location in the HDV genome sequence. SOURCE: ACCESS Health International (Adapted from: Wille et al. 2018 https://doi.org/10.3390/v10120720)
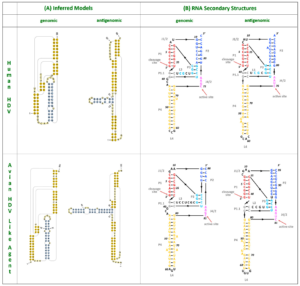
FIGURE 2. HDV ribozymes. (A) Secondary structures of the genomic and antigenomic ribozymes inferred using the TT2NE algorithm. The HDV ribozyme models were used as reference to screen for the ribozyme sequences in the avian HDV-like genome sequence. (B) Re-drawn secondary structures of the genomic and antigenomic ribozymes. SOURCE: Wille et al. 2018
Not Just Ducks, Toads and Termites Too
Spurred on by these findings, Wille and colleagues revisited data collected during previous metatranscriptomic work, now with an eye towards identifying potential new hepatitis D virus-like agents. This earlier data included samples from amphibians, fish, reptiles, and invertebrates.
From the more than a billion nucleotides gathered across the various libraries, the researchers were able to isolate four novel hepatitis D virus-like agents. These belonged to Subterranean termite, Asiatic toad, Chinese fire belly newt, and a mixture of fish (Table 1).
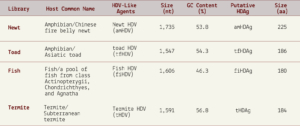
TABLE 1. Characterization of the HDV-like agents and their putative HDAgs. SOURCE: ACCESS Health International (Adapted from: Chang et al. 2019 https://doi.org/10.1093/ve/vez021)
As with the duck hepatitis D virus-like agents, the guanine/cytosine contents of these newly discovered varieties were also lower than in human hepatitis D virus. Still, all of the genomes were circular with unbranched rod-like secondary structures and their sizes consistent with other hepatitis D virus-like agents (1500-1700 nucleotides). Additionally, all of the newly discovered agents encoded a putative hepatitis D virus-like antigen protein, which, when cross-referenced against large protein databases, proved to be most closely related to the human delta antigen proteins. Even for the distinct domains within each of the putative antigen proteins, all of the “top scoring hits” matched human hepatitis D antigen protein.
Again, none of the data libraries contained any hepatitis B virus RNA. In fact, they didn’t contain any hepadnavirus RNA of any kind. They did, however, contain sequences from other viruses which may have served a functionally similar role (Table 2).

TABLE 2. Details of the HDV-like agents and associated viruses discovered previously. SOURCE: ACCESS Health International (Adapted from: Chang et al. 2019)\
This adds credence to the notion that hepatitis D virus and virus-like agents are not exclusively tethered to the hepatitis B virus and may instead be able to use other helper viruses to assist with transmission and infection.
Implications
Since the completion of these early studies, the list of potential hosts has been growing steadily. Aside from ducks, newts, toads and the rest of the abovementioned animals, hepatitis D virus-like agents have now also been found in white-tailed deer, passerine birds, and woodchucks. Clearly, the assumption that hepatitis D virus is a strictly human pathogen has reached its limit — the virus is far more widespread than initially thought. Not only this, but the range of helper viruses is also likely far more diverse than previously understood. None of the animal samples showed any indication of co-infection with hepatitis B virus, and yet hepatitis D virus persisted in great numbers. We have to begin acknowledging that hepatitis D virus-like agents likely adopt envelope proteins from a variety of sources. Different helper viruses may enable the infection of different tissues, and with this, cause diseases never previously associated with hepatitis D virus. We are only just beginning to scratch the surface.

