Does The Hepatitis D Genome Encode Proteins From Both The “Plus” And “Minus” Strands?
(Posted on Tuesday, May 30, 2023)
This story is part of a larger series on viroids and virusoids, small infectious RNAs. It is also the fourth installment in a series on hepatitis D virus, a virusoid-like pathogen that causes serious human disease. You may read the others on Forbes or www.williamhaseltine.com. Here, we take a look at a large open reading frame (ORF) in the positive strand of the virus.
Viral hepatitis is a disease characterized by inflammation of the liver. Over time, this inflammation can lead to severe and often-irreversible scarring, interfering with the liver’s ability to perform its necessary functions. At its most extreme, the spread of scar tissue causes complete failure of the liver.
Although there are multiple hepatitis viruses, all of which cause disease, one of them is particularly pernicious: hepatitis D virus. Despite the name, hepatitis D virus more closely resembles a class of subviral pathogens called virusoids — small, circular RNAs that lack a protective protein coat and depend on a helper virus to transmit between hosts. In cases of human infection, hepatitis D is usually carried in by hepatitis B virus. The latter lends hepatitis D a protective coat replete with surface proteins, which enable entry into host cells.
Somewhat unusual for virusoids, hepatitis D produces two proteins. These are encoded by a stretch of RNA called an open reading frame (ORF); in brief, this is a section of the genome that can be “read” by host ribosomes, special cellular machines that translate RNA into proteins. A genome can contain multiple such open reading frames, each of which might encode a different protein. Indeed, aside from the open reading frame that encodes its two antigen proteins, hepatitis D virus contains a few additional open reading frames. Discussion of these open reading frames, and their potential protein-producing capacity, is oddly absent from the literature. What follows is an attempt to fill this gap by providing a preliminary analysis of the additional open reading frames.
Genome and Antigenome
Every RNA genome is made up of an elaborate mix of four basic building blocks, called nucleotides, which are defined according to their nitrogenous base: adenine (A), guanine (G), cytosine (C), or uracil (U). Each base has its complementary pair; Adenine links up with uracil, and guanine links up with cytosine.
As part of the hepatitis D virus replication process, the RNA genome is used as a template for the production of a complementary, antigenomic copy (Figure 1). This copy is the exact inverse of the original genome — a mirror image. Wherever there’s an adenine in the genome, the antigenome instead contains a uracil, and so on. The genome is also referred to as the “plus” strand, and the antigenome as the “minus” strand.
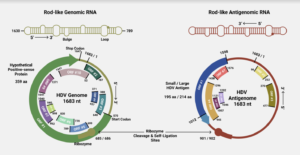
FIGURE 1. (Top) Schematic of the rod-like shape of the hepatitis D virus (HDV) genome/antigenome. (Bottom) Overview of the genome and antigenome of HDV, with ribozyme cleavage sites and open reading frames (ORFs) clearly marked. SOURCE: ACCESS Health International (Adapted from: Magnius et al. 2018, https://doi.org/10.1099/jgv.0.001150)
In the case of hepatitis D, the open reading frame that encodes the two antigen proteins is located in the antigenome. But the genomic RNA also contains open reading frames — of course, there is plenty of variation from strain to strain, with some containing longer open reading frames than others. Occasionally, you’ll find a strain with a very large open reading frame. Indeed, one strain contains an open reading frame in the genomic RNA that clocks in at 359 amino-acids-long. Composed of 1077 nucleotides, it spans almost two thirds of the entire genome. Compare this to the antigen-encoding open reading frame of the antigenomic strand, which is 214 amino-acids-long.
Whether or not the open reading frames found in the genomic RNA of hepatitis D actually produce any proteins is unclear; all the pieces are in place, but that doesn’t necessarily mean they are being put to use. For example, there is no evidence at present that a messenger RNA corresponding to the positive strand is made, still less that it is transported to the cytoplasm for translation. Perhaps it is only present at some stage of the life cycle and disease process.
Still, it seems extremely unlikely that such a long open reading frame would be present and yet remain unused. This is especially true as the constraints to maintain such a long open reading frame overlapping the minus strand coding sequence is severe. This observation calls for an intense search for both positive strand messenger RNAs and proteins.
In addition to the long open reading frame, there are also a number of other, shorter open reading frames scattered throughout the genome. This is also true of the antigenome, which aside from the antigen-producing open reading frame, has multiple other open reading frames that could potentially encode proteins. Whether or not any of these additional open reading frames are functional remains to be seen.
We recommend that a serious effort be mounted to test for the presence of the hypothetical proteins of the genomic-strand RNA. This can be achieved by creating antibodies that target the hypothetical protein. Testing should be done not only for the entire replication cycle of hepatitis D virus, but for all stages of infection and disease progression.
Functions?
Assuming the aforementioned 359 amino-acids-long open reading frame did encode a protein, what functions might it have? By comparing the amino acid sequence of the open reading frame to sequences listed in the GenBank database, we were able to identify matches to three proteins: phosphohistidine phosphatase, GDP-L-fucose synthase, and pectate lyase.
It is worth noting that these matches are mainly partial matches, meaning only a small section of the open reading frame overlaps with a small section of the protein sequence. Extrapolating what function a particular snippet of a protein sequence might have is an inexact science; the regions of similarity may have particular functions that do not necessarily match the overall functions after which the full proteins are named. What follows should be understood as laying the initial groundwork for further, more detailed research.
It is also important to keep in mind that the open reading frame discussed above is found in one strain of hepatitis D virus, but how conserved it is across strains is unclear. It would be prudent to first establish whether there are any open reading frames in the genomic RNA that are highly conserved, and then use these highly conserved sequences for future analysis.
Phosphohistidine Phosphatase
Phosphohistidine phosphatase is a protein involved in “dephosphorylation”, the removal of a phosphate group from a molecule. Phosphorylation and dephosphorylation are crucial processes in the regulation of cell signaling and in the regulation of gene expression. The addition or removal of a phosphate group can contribute to whether a certain gene, and by extension protein, is expressed or suppressed.
It is not clear exactly how this would influence the hepatitis D life cycle, but considering the importance of de/phosphorylation to virtually every cellular process —including transcription and translation— it could be an important part of the puzzle.
GDP-L-Fucose Synthase
As implied by its name, GDP-L-fucose synthase is a protein that catalyzes the production of GDP-L-fucose. GDP-L-fucose, in turn, is a critical player in a process known as fucosylation, which is characterized by the addition of fucose sugar units to molecules. Fucosylation of proteins is relevant to both cell signaling and immune regulation.
That said, fucosylation has also been associated with various cancers, including liver cancer. It is well known that chronic hepatitis D infection can lead to liver cancer, and it does so more frequently than hepatitis B monoinfection. The mechanisms underlying this increased risk for liver cancer are poorly understood. Perhaps the open reading frames of the hepatitis D genomic RNA contribute to disease state, rather than replication and life cycle? Additional research is needed to confirm whether this may be the case.
Pectate Lyase
Pectate lyases are enzymes involved in the degradation of plant cell walls. Although employed in certain plant tissues to help remodel the cell wall during growth, they are most commonly found in plant pathogens, including bacteria. Here, they help the pathogens break down pectin, which plays a key role in the rigidity of the cell wall; think of a battering ram breaking down a castle gate. Once the cell wall is weakened, entry becomes easy.
The use of pectate lyase-like functions to hepatitis D is unclear. Viroids and most virusoids, with which hepatitis D shares many similarities, are plant pathogens. Yet, they rarely encode any of their own proteins. This makes it unlikely that the similarities between the large genomic-strand open reading frame and pectate lyase are remnants of a shared evolutionary history between hepatitis D and viroids.
Since hepatitis D, per current understanding, exclusively infects non-plant hosts, it seems unlikely that the pectate lyase-like sequence of the open reading frame contributes heavily to the viral life cycle.
Implications