Progress In The Search For Broadly Neutralizing Monoclonal Antibodies IV
(Posted on Thursday, July 28, 2022)
This is part of a continuing series describing antiviral antibodies to prevent and treat SARS-CoV-2 infections. In this series, we will discuss the fundamental nature of virus evolution, how SARS-CoV-2 has mutated to evade neutralizing antibodies, and our latest attempts to fight against these mutations with more recent and improved antibody candidates.
Monoclonal antibody treatments remain our greatest asset in the fight against Covid-19, though in recent months, the effectiveness of these treatments has waned. New variants with more mutations have evolved to overcome antibodies from monoclonal therapies, vaccines, and prior infections. The search for monoclonal antibodies that neutralize not one but all strains is underway to counter these new variants.
In this series, we have discussed several pan-variant monoclonal antibodies, all of which promise against current Omicron strains and previous variants of concern such as Alpha, Beta, and Delta. Here we analyze another described in a study by Zhou et al.: the ZCB11 antibody.
ZBC11 Antibody Origin
Hong Kong researchers used an uncommon antibody identification method in their study. Antibodies described previously in this series were identified either from the sera of those naturally infected with Covid-19 or from a specially-engineered mouse model. Here researchers use the sera of mRNA vaccine inoculated patients as the basis for antibody collection.
Collecting the sera of 34 Pfizer vaccinated subjects, Zhou et al. found that only two of the 34 samples had neutralizing activities against all variants of concern included in the study, notably Alpha, Beta, Gamma, Delta, and Omicron BA.1. Subject #26 displayed the highest neutralization titers against Beta and Delta. They showed above-average neutralization of Alpha, Gamma, and Omicron. Zhou et al., therefore, took a closer look at the sera of this vaccinee for broadly neutralizing antibodies.
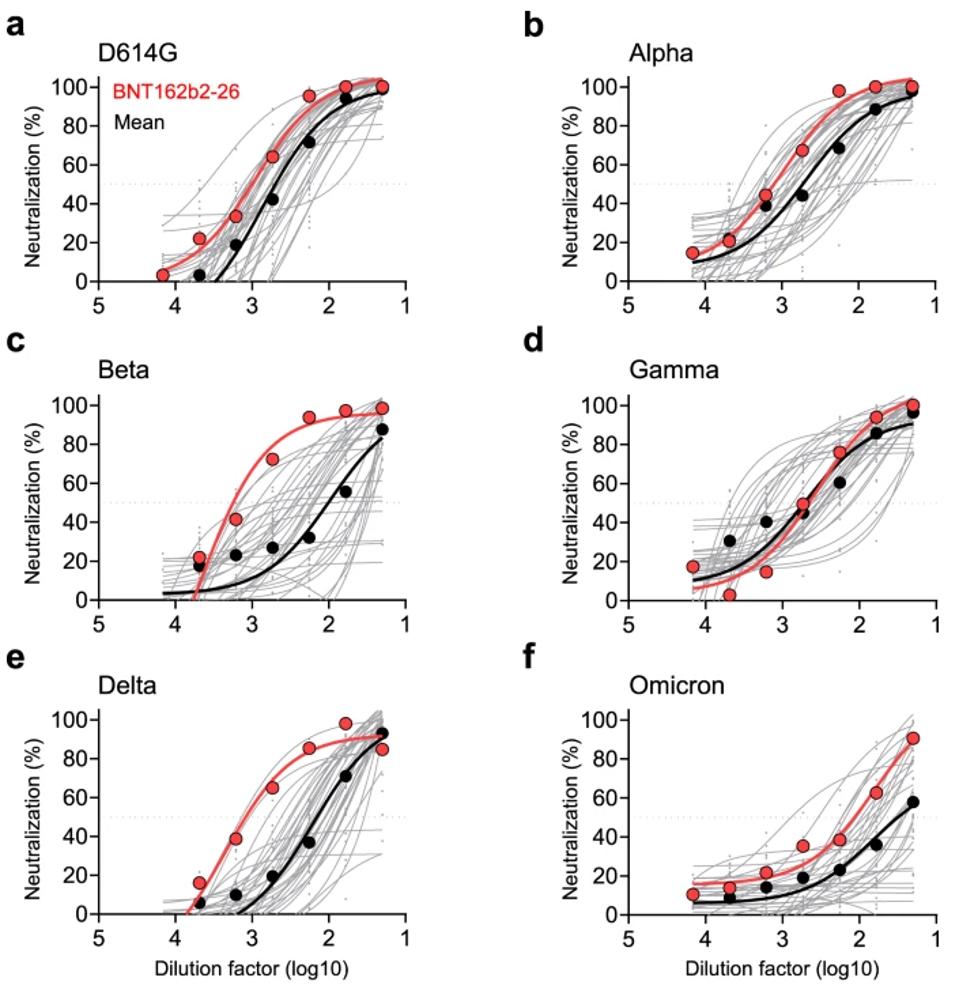
FIGURE 1: Serially diluted plasma samples subjected to neutralization assay against the pseudotyped SARS-CoV-2 WT (a) and five VOCs (b–f).
ZHOU ET AL.
The researchers collected another blood sample 130 days after the second vaccination. Fourteen antibodies were identified, while only seven (ZCB3, ZCB8, ZCB9, ZCB11, ZCC10, ZCD3, ZCD4) showed positive responses to WT spike. They narrowed these seven to four by limiting their search to receptor-binding domain-specific antibodies that displayed neutralization against the wildtype Wuhan strain, namely ZCB3, ZCB11, ZCC10, and ZCD3. Along with a control antibody, ZB8, these four moved onto VOC neutralization assays.
ZCB11 Antibody Neutralization
Zhou et al. conducted both pseudovirus and live virus assays for the four antibody candidates. Against the Alpha, Beta, Gamma, Delta, and Omicron BA.1 pseudotypes, ZCB11 was the only candidate to neutralize all variants and consistently outperformed other antibodies when they did neutralize.
Against authentic viruses, this result was replicated to an even greater degree. ZCB11 consistently and effectively neutralized all variants in the lineup, including Omicron BA.1, BA.1.1, and BA.2. We note that the currently circulating BA.4 and BA.5 were not included in the neutralization assays. Still, the potent neutralization of BA.2 and all earlier strains strongly indicates ZCB11’s broad neutralization.

FIGURE 2: Neutralization IC50 values of NAbs against the wildtype, Alpha, Beta, Gamma, Delta, and Omicron strains. The lower the IC50 values, the greater the neutralization.
ZHOU ET AL.
ZCB11 Antibody Target
Alongside many other monoclonal antibodies, The ZCB11 antibody targets the receptor-binding motif. Cyro-electron microscopy analysis of the antibody revealed the antibody binding to the Spike protein in the “up” conformation, wherein the Spike is preparing to bind the ACE2 receptor of the host cell.
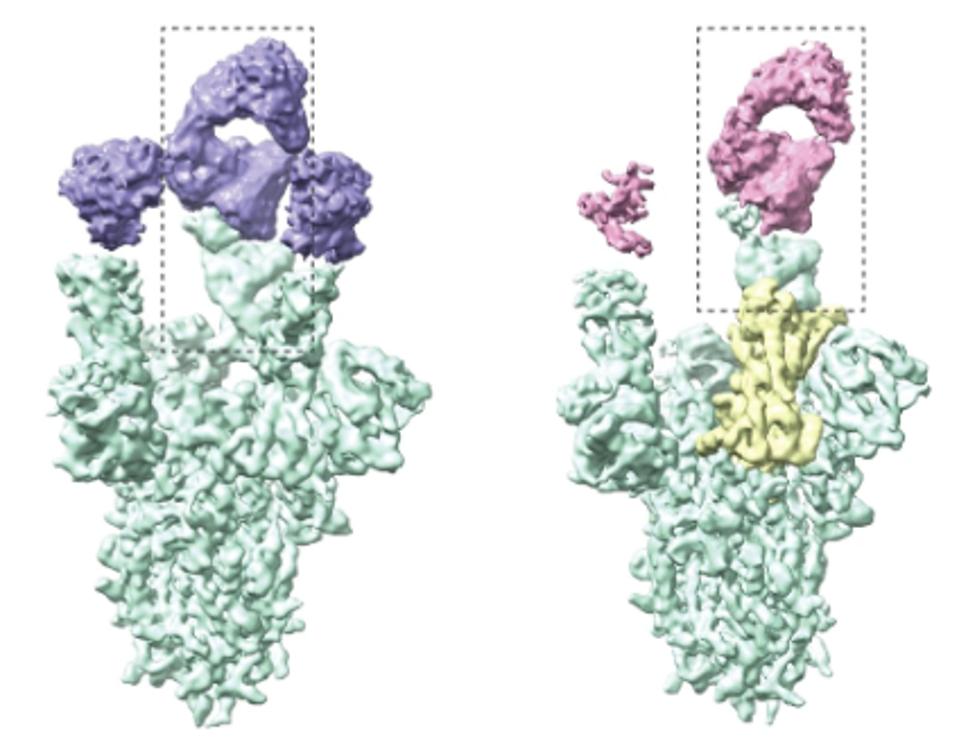
FIGURE 3: Cryo-EM density map of spike trimer in complex with ZCB11 Fab. Two of three different states (3 u and 2u1d) are shown. Spike trimer is color-coded in green and Fab is in pink and purple in two states, respectively. Down RBD is color-coded i
ZHOU ET AL.
Why does this antibody neutralize much more effectively than previous monoclonal treatments when they all bind the receptor-binding domain? We can attribute this to two things. One reason for the broad activity found with ZCB11 is that most of the amino acid contacts are highly conserved among all known coronavirus sequences.
The antibody footprint binds the Spike amino acids D420, L455, F456, N460, A475, S477, T478, F486, and N487. Three of these, S477, T478, and F486 are mutated in the latest Omicron strains to S477N, T478K, and F486V. In the GISAID SARS-CoV-2 sequence database, all positions aside from the three Omicron exceptions are mutated less than 5,000 times in over 12 million sequences, meaning the footprint of this antibody is highly conserved.
Notably, Zhou et al. found that S477N and T478K increase the binding affinity between the antibody and Spike, rather than interfering with neutralizing capability. They postulate that this is at least a partial explanation for the antibody’s substantial neutralization of the variant.
The second reason is that most of the amino acid positions in ZCB11 are relatively unmutated in natural strains. Previous antibodies have contact points at major residues of mutation like N501 and E484, but ZCB11 mostly lacks contact points at major mutational residues.
As we recommend with all broadly neutralizing antibodies, there is no reason to limit treatment to just one. An antibody cocktail of two or three monoclonal antibodies covering a broad footprint of conserved residues could be a powerful weapon against current and future strains, which are sure to continue mutating to evade immunity. We must prioritize and expedite these antibodies’ production as the pandemic continues to rage.