The Many Functions Of The Hepatitis D Antigen Proteins
(Posted on Tuesday, May 23, 2023)
This story is part of a larger series on viroids and virusoids, small infectious RNAs. It is also the third installment in a series on hepatitis D virus, a virusoid-like pathogen that causes serious human disease. You may read the others on Forbes or www.williamhaseltine.com. Here, we take a look at the proteins encoded by hepatitis D virus.
Hepatitis D virus is a small circular satellite RNA that can cause serious disease — despite its tiny size, it is responsible for the most severe form of viral hepatitis. Being a satellite RNA, it depends on a helper virus to infect cells and transmit between hosts. In humans, the usual accomplice is hepatitis B virus, which “lends” hepatitis D its protein coat, including surface proteins. Once inside a host cell, however, the two viruses go their separate ways and hepatitis D replicates independently. Vital to this replication process are the two proteins that hepatitis D virus produces: the small antigen and the large antigen.
Indeed, the hepatitis D antigens was the first clue in the mystery that ultimately led to the discovery of hepatitis D virus. In 1977, a gastroenterologist by the name of Mario Rizzetto, working out of the University of Turin, Italy, noticed some unusual activity in liver biopsies of chronically ill hepatitis B patients: some samples showed antibody activity against the hepatitis B core antigen even when there was none present. After a series of follow-up experiments, Rizzetto and his colleagues realized that they had happened upon the antigen protein of an entirely different hepatitis virus. They named the new pathogen δ, delta. Later work revealed the presence of another, larger antigen, now called the large hepatitis D antigen. What follows is a detailed overview of these two proteins and their role in the hepatitis D virus life cycle.
Proteins At a Glance
As mentioned, the hepatitis D virus encodes two different proteins from the same open reading frame (ORF): the small hepatitis D antigen (S-HDAg), 195 amino acids, and the large hepatitis D antigen (L-HDAg), 214 amino acids. The open reading frame that encodes the proteins is found in the hepatitis D antigenomic strand. The genomic strand also contains a large open reading frame, but whether it actively encodes any proteins has yet to be addressed in the current literature.
The difference between the two antigen proteins is a 19-amino-acid extension at the end of the small antigen (Figure 1). Despite sharing the majority of their amino acids, the functions of these two proteins within the hepatitis D life cycle are vastly different.
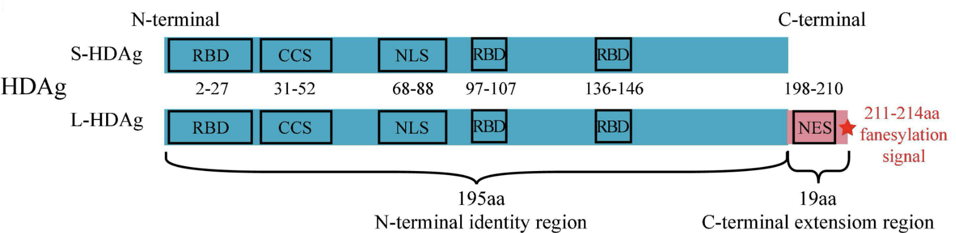
FIGURE 1. S-HDAg and L-HDAg are characterized by three RNA-binding domains (RBD), a coiled-coil sequence (CCS), a nuclear localization signal (NLS) in the N-terminal identity region (blue box), and a nuclear export signal (NES), a farnesylation signal (red star) in the C-terminal extension region (pink box). SOURCE: Zi et al. 2022 https://doi.org/10.3389/fmicb.2022.838382
Beads on a String: Ribonucleoprotein Complex
The antigens produced by the hepatitis D virus are integral to replication. They are also integral to the structure of the complete “viral” particle. Eight single units of the small hepatitis D antigen bind together to form one big entity, known as an octamer. The same happens with the large hepatitis D antigen. Some of the octamers are also formed via a mix-match of the large and small antigens. Four or five of these octamers are then grouped together and bound by the hepatitis D virus RNA. Based on the amount of hepatitis D virus antigens present per viral particle —roughly between 70 and 200— it is likely that somewhere between two to six such bundles get wrapped up by the RNA. The finished product, known as a ribonucleoprotein complex, resembles beads on a string (Figure 2).
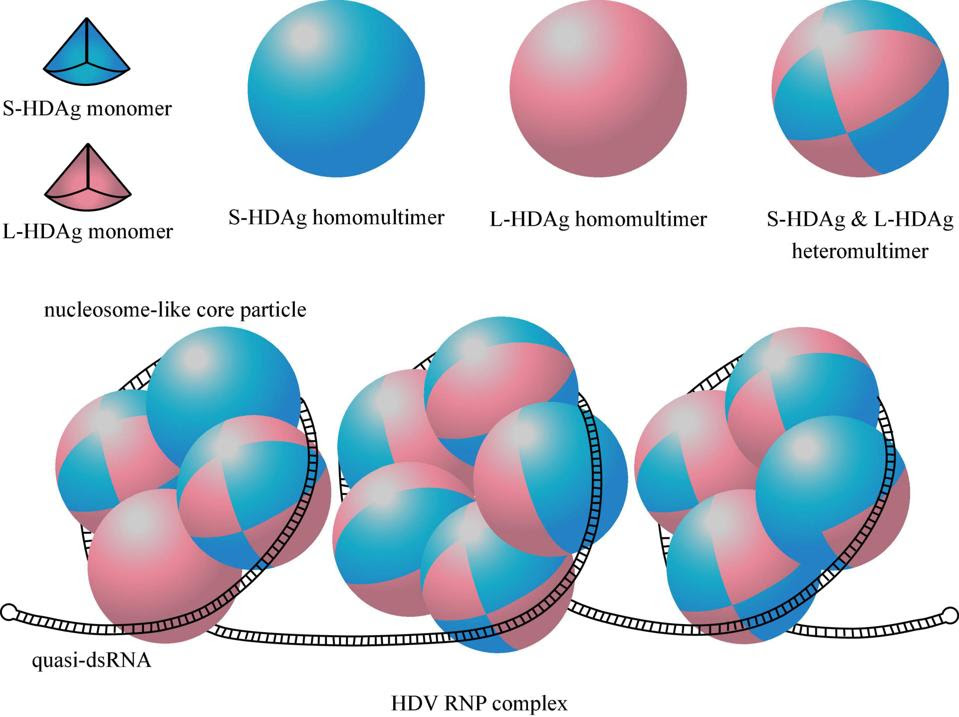
FIGURE 2. Structure of HDAg multimers and HDV ribonucleoprotein (RNP) complex. One eighth of the sphere represents a HDAg monomer, the blue one represents S-HDAg, the pink one represents L-HDAg. A HDAg octamer containing 8 S-HDAg or 8 L-HDAg is called homomultimer, and is called heteromultimer when it contains a mix of S-HDAg and L-HDAg. Quasi-double-stranded RNA (quasi-dsRNA) wraps around four or five HDAg octamers as a nucleosome-like core particle, and the whole HDV RNP contains 2–6 such core particles. SOURCE: Zi et al. 2022 https://doi.org/10.3389/fmicb.2022.838382
Finally, the whole ribonucleoprotein complex is encapsulated by a protein coat dotted with the hepatitis B virus surface antigen. This protein coat protects the genetic material of the hepatitis D virus and enables entry into new host cells. The hepatitis B antigen protein comes in three versions: a small version, a medium version, and a large version. All three can be found in the protein coat surrounding the hepatitis D virus genome.
Small Hepatitis D Antigen
The first 195 amino acids, corresponding to the small hepatitis D antigen, contain three crucial functional domains. The RNA-binding domain (RBD) allows the antigen to bind to the hepatitis D virus genome and form a ribonucleoprotein complex. There are three RNA-binding-domain regions in the antigen: amino acids two to 27, 97 to 107, and 136 to 146. The next crucial domain is the “coiled-coil” sequence (CCS), which spans from amino acid 31 to amino acid 52. This helps mediate the polymerization of the antigen. Finally, amino acids 68 to 88 represent the nuclear localization signal (NLS), which serves to catalyze movement of the antigen from one part of the cell to another; from the cytoplasm to the nucleus.
When hepatitis D virus first enters a host cell, the nuclear localization signal in the small antigen protein ferries it to the nucleus. There, the virus appropriates a host enzyme, RNA polymerase II (Pol-II), to create multiple copies of its messenger RNA sequence. Traditionally, host cells use this enzyme to transcribe DNA into RNA, but hepatitis D virus manages to hijack it to reproduce RNA from RNA. Although it is still not fully understood how this happens, researchers suspect it has to do with the quasi-double-stranded structure of the hepatitis D virus genome, which the cell enzyme might mistake for DNA.
The mRNAs exit the nucleus and travel into the cytoplasm, where they are translated into small hepatitis D virus antigens (Figure 3). These antigens then migrate back into the nucleus, ready to assist with replication.
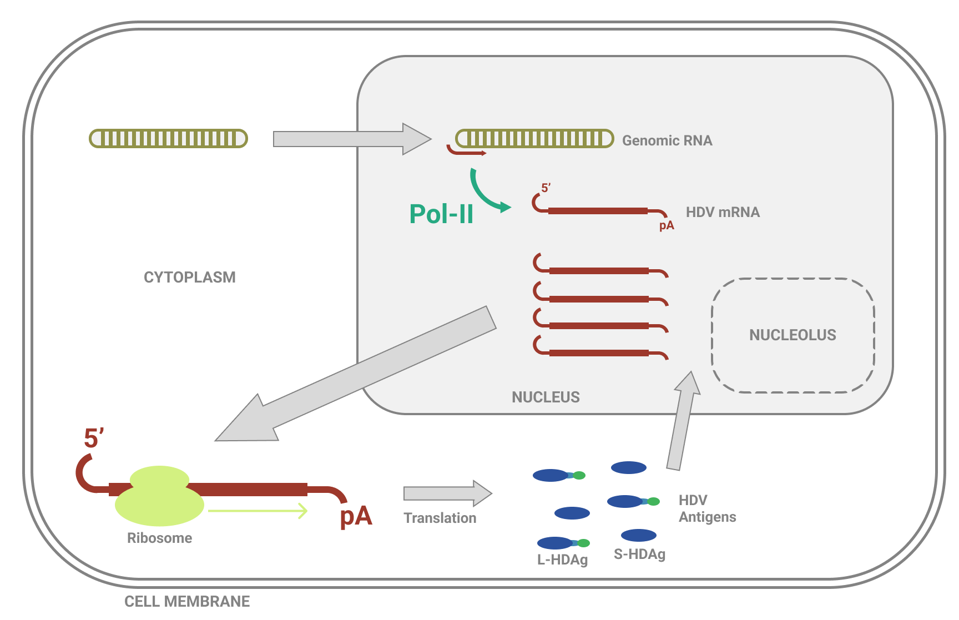
FIGURE 3. Overview of hepatitis D virus (HDV) mRNA synthesis. Abbreviations: 5’, five-prime end; pA, poly-A tail; Pol-II, DNA-dependent RNA-polymerase II; L-HDAg, large hepatitis D antigen; S-HDAg, small hepatitis D antigen. SOURCE: ACCESS HEALTH INTERNATIONAL
Once in the nucleus, the newly synthesized small antigens bind to RNA polymerase II. In doing so, the antigens displace a protein complex called negative elongation factor (NELF), which usually acts as a gatekeeper of transcription. Having cleared the way, amplification of hepatitis D genomic RNA can begin. By binding to RNA polymerase II, the small antigens also loosen the grip that the polymerase “clamp” —think of this as a hand that allows the polymerase to latch onto DNA or RNA— has on the hepatitis D virus RNA. Loosening the grip that the polymerase has on the RNA allows it to move along more quickly than it usually would, accelerating the transcription process.
The small hepatitis D antigens also bind, via amino acids two to 67, to the central globular domain of human histone 1E (H1E). Histones are proteins in the nucleus which bind to and wrap up long strands of DNA, compacting them into structures called chromosomes. They also help regulate gene activity. Exactly how this helps the replication process of hepatitis D is unknown, but it is crucial: eliminating the hepatitis D binding site on histones completely inhibits replication.
Aside from RNA polymerase II and histone 1E, small hepatitis D antigens interact with Yin Yang 1. This is a transcription factor, meaning it plays a role in regulating the speed of transcription. The RNA-binding domain of the small hepatitis D antigen binds to part of Yin Yang 1 as well as two enzymes associated with Yin Yang 1, CREB-binding protein (CRB) and p300. The resulting protein complex helps enhance RNA amplification, accelerating the hepatitis D replication process.
Finally, small hepatitis D antigens also bind to a chromatin remodeling protein called bromodomain adjacent to zinc finger domain 2B (BAZ2B). Chromatin is a DNA-protein complex that condenses the long strands of DNA into more compacted structures — if our DNA wasn’t wrapped up like this, it would be far too long to fit in our cells. Chromatin remodeling proteins influence gene expression by modulating the chromatin architecture. Although it is not completely clear how, hepatitis D interaction with the bromodomain adjacent to zinc finger domain 2B protein is critical to sustained RNA replication. Interfering with the interaction between the two inhibits RNA amplification.
Large Hepatitis D Antigen
Whereas the small hepatitis D antigen kickstarts and accelerates replication of hepatitis D RNA, the large hepatitis D antigen acts as a kill switch, inhibiting further replication and instead facilitating a shift towards viral assembly.
To this end, the 19-amino-acids-long extension of the large hepatitis D antigen contains a nuclear export signal (NES), which helps shuttle the ribonucleoprotein complex from the nucleus of the host cell into the cytoplasm, where it can be wrapped up in its protein coat. The last four amino acids of the large hepatitis D antigen make up a farnesylation signal. This signal recruits a host protease, farnesyl transferase, to add a small molecule called a farnesyl lipid group to the end of the large antigen. The addition of the farnesyl lipid group is critical to viral assembly, as it acts like a “hook” that enables the hepatitis D ribonucleoprotein complex to combine with the hepatitis-B-antigen-containing envelope (Figure 4). Only once it has been enveloped can the hepatitis D virus particle go on to infect new cells and transmit to new hosts.
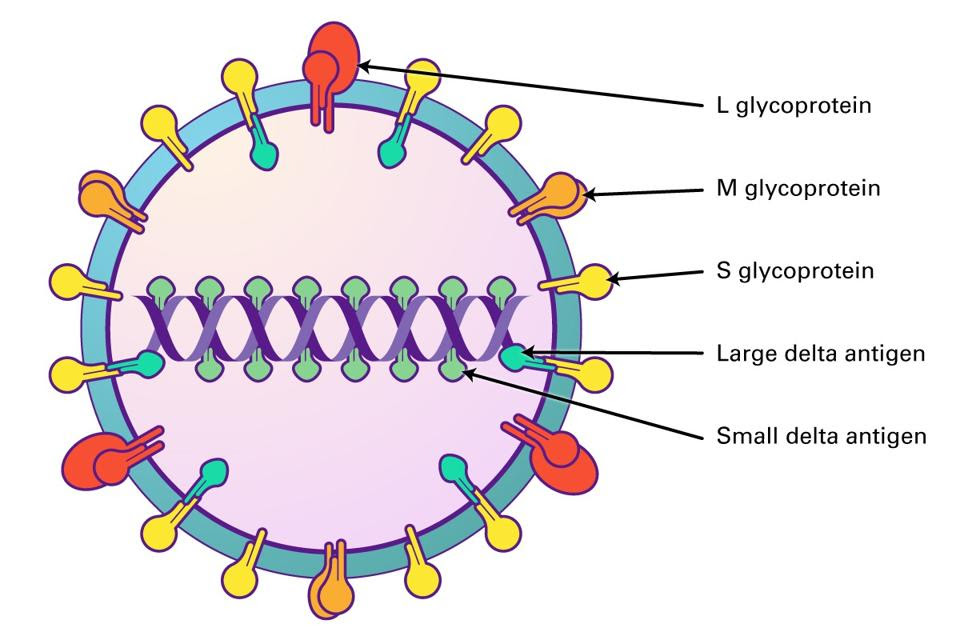
FIGURE 4. The farnesyl lipid group at the end of the large delta antigen (L-HDAg) latches onto hepatitis B surface antigens and creates a protective coat protein. SOURCE: LIFESPAN BIOSCIENCES
Large Hepatitis D Antigen Production
Since the same stretch of RNA encodes both the small and large hepatitis D antigens, there has to be some mechanism that shifts production from the small antigen to the large antigen at just the right time. Here, hepatitis D virus employs a nifty bit of sleight of hand.
To transition from production of the small delta antigen to the large delta antigen, a single mutation of the mRNA sequence takes place, catalyzed by a protein called the host adenosine deaminase acting on RNA 1 (ADAR1). After a sufficient amount of small hepatitis D antigens have gathered in the nucleus, adenosine deaminase acting on RNA 1 binds to the hepatitis D RNA and changes the UAG stop codon, which usually marks the end of the open reading frame, to UGG. This mutation extends the open reading frame by 57 nucleotides, and thus adds a 19-amino-acids-long extension to the end of the small delta antigen (Figure 5).
Curiously, this point mutation doesn’t occur directly on the hepatitis D antigen mRNA, but rather on the full-length antigenomic strand used as an intermediate during RNA replication. As the antigenomic strand is transcribed back into its genomic counterpart, the mutation carries over. The mRNA sequences derived from this mutated hepatitis D genome will from then on produce the 214-amino-acid large delta antigen.
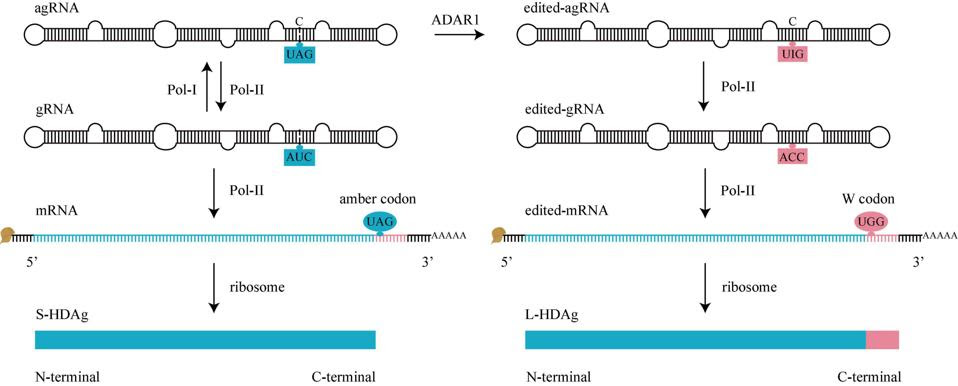
FIGURE 5. HDV RNA editing. Before editing (left), there is an amber codon (UAG, blue oval) at amber/W site of mRNA, translation will stop here to produce S-HDAg. During editing, adenosine deaminases acting on RNA 1 (ADAR1) converts UAG of agRNA to UIG (right). When Pol-II use these edited-agRNA as a template to replicate edited-gRNA, and use edited-gRNA as a template to transcribe edited-mRNA, there is a W codon (UGG, pink oval) at amber/W site of edited-mRNA, the translation won’t stop until next termination codon is met, producing an L-HDAg. gRNA, genomic RNA. agRNA, antigenomic RNA. SOURCE: Zi et al. 2022
Over time, the large hepatitis D antigen catches up to the small hepatitis D antigen, and the proportion of the two antigens in the ribonucleoprotein complex equalizes. After equilibrium of the two antigen proteins has been reached, the nuclear export signal of the large hepatitis D antigen becomes the dominant message. This acts as a trigger for migration of the ribonucleoprotein complex out of the nucleus and into the cytoplasm, where it can be wrapped up in a protein coat before finally exiting the cell.