What Are The 677 Mutations? New Covid-19 Variants Found In The US
(Posted on Friday, February 19, 2021)
Random variation is an essential component of all living things. It drives diversity, and it is why there are so many different species. Viruses are no exception. Most viruses are experts at changing genomes to adapt to their environment. We now have evidence that the virus that causes Covid, SARS-CoV-2, not only changes, but changes in ways that are significant. This is the sixteenth part of a series of articles on how the virus changes and what that means for humanity. Read the rest: part one, part two, part three, part four, part five, part six, part seven, part eight, part nine, part ten, part eleven, part twelve, part thirteen, part fourteen, and part fifteen.
Colorized scanning electron micrograph of a cell showing morphological signs of apoptosis, infected with SARS-COV-2 virus particles (orange), isolated from a patient sample. Image captured at the NIAID Integrated Research Facility (IRF) in Fort
BSIP/UNIVERSAL IMAGES GROUP VIA GETTY IMAGES
This winter, new variants of SARS-CoV-2 have taken the world by storm. They have emerged in corners of the globe as disparate as Brazil and the United Kingdom; exhibited an array of new and worrisome properties, ranging from increased transmissibility to immunogenicity; and complicated our best efforts to collectively mitigate the spread of Covid-19 through public health measures and vaccines. Earlier this week I wrote a piece detailing why this trend is so concerning for the United States, where variants of concern have been identified in states like Ohio and California but otherwise remain relatively under the radar, circulating unbeknownst to most Americans. In today’s installment, I want to focus on the recent appearance of what researchers are calling the 677 variants, a crop of lineages that at least for now seems confined to the US alone.
A hallmark of viral variation is that the tiny alterations distinguishing one variant from the next can arise in separate locations at the same time, developing independently of one another. One example is the N501Y spike protein mutation that is known to increase the affinity of the virus for our ACE2 receptors. N501Y was first spotted in the genome of the B.1.1.7 (UK) variant, but it didn’t take long for scientists to discover its presence in the B.1.351 (South Africa) and P.1 (Brazil) variants as well. Another spike protein mutation linked to immune escape, E484K, has also acquired high global frequency, appearing first in B.1.351 and more recently in B.1.525, the latest variant to wash up on British shores.
The 677 variants, according to a preprint study published last week, are similar in that they were first detected in October by genomic surveillance programs nearly a thousand miles apart. They’re named for their one commonality—mutations at the 677 amino acid site. In Louisiana, where one of the programs to successfully trace them is located, researchers found that the proportion of SARS-CoV-2 viruses in local circulation carrying a Q677P mutation rose from nil to nearly a third (28 percent) between early December 2020 and late January 2021. Within the same timeframe, the mutation’s prevalence in New Mexico, where the other program is based, shot up to just over 11 percent.
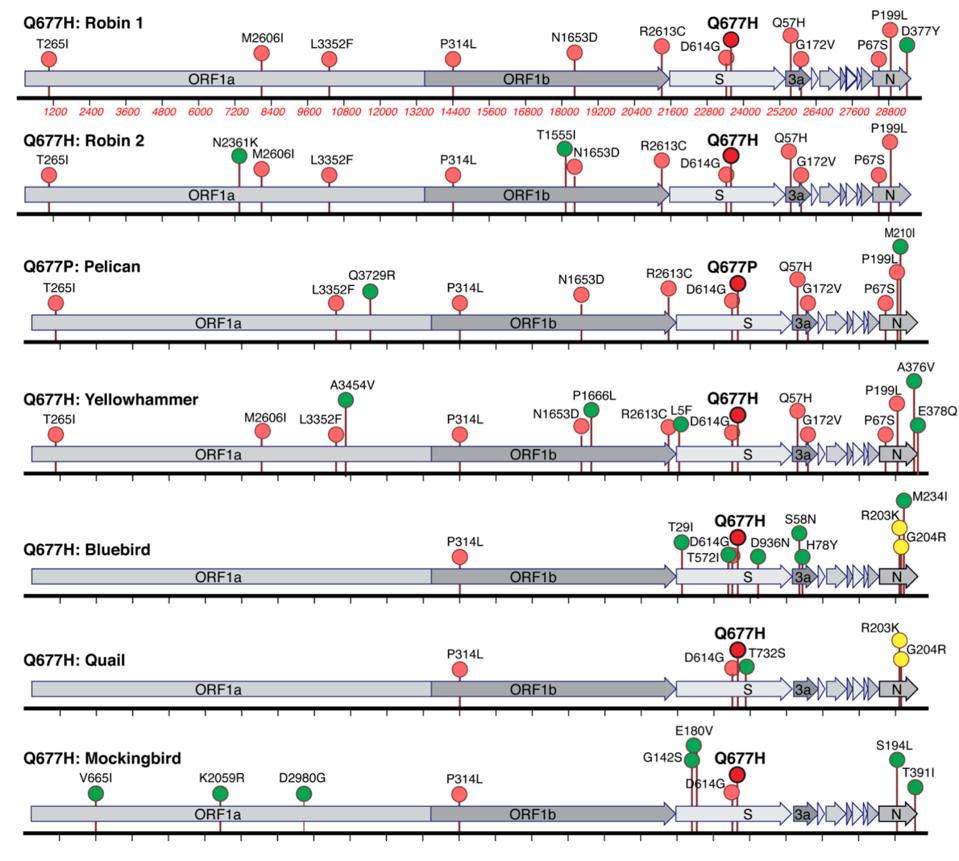
Figure 1. Lineage-specific and shared amino acid polymorphisms of US S:Q677P and S:677H variants.
SOURCE: “EMERGENCE IN LATE 2020 OF MULTIPLE LINEAGES OF SARS-COV-2 SPIKE PROTEIN VARIANTS AFFECTING AMINO ACID POSITION 677” HTTPS://WWW.MEDRXIV.ORG/CONTENT/10.1101/2021.02.12.21251658V2
The researchers were so intrigued by the convergence around the 677 site that they decided to seek out other 677 point mutations by conducting phylogenetic analysis on available genomic data (Figure 1). One mutation, Q77H, appeared in six distinct lineages from August to November 2020. Though four of the six were confirmed in only about 100 Covid-19 cases or fewer, the remaining two popped up with considerably more frequency—one with nearly 300 cases and the other close to 800. Slight as these figures may seem compared to the 28 million cases reported in the US so far, given the relative paucity of US genomic sequencing efforts they do indicate a potential for further spread.
I’ve written previously about a variant spreading rapidly in Columbus, Ohio that has the Q77H mutation, though in that particular state the UK B.1.1.7 variant is reportedly disposed to become the dominant stain by March or April, according to Dr. Bruce Vanderhoff, their chief medical officer. The emergent B.1.525 variant, which has been identified in Nigeria, Denmark, the UK, and several other countries, also carries the Q77H mutation, making it the only international variant to do so. But beyond that, mutations on the 677 site are conspicuously absent from all the variants that have become dominant around the world thus far, appearing almost exclusively in nascent lineages in the US.
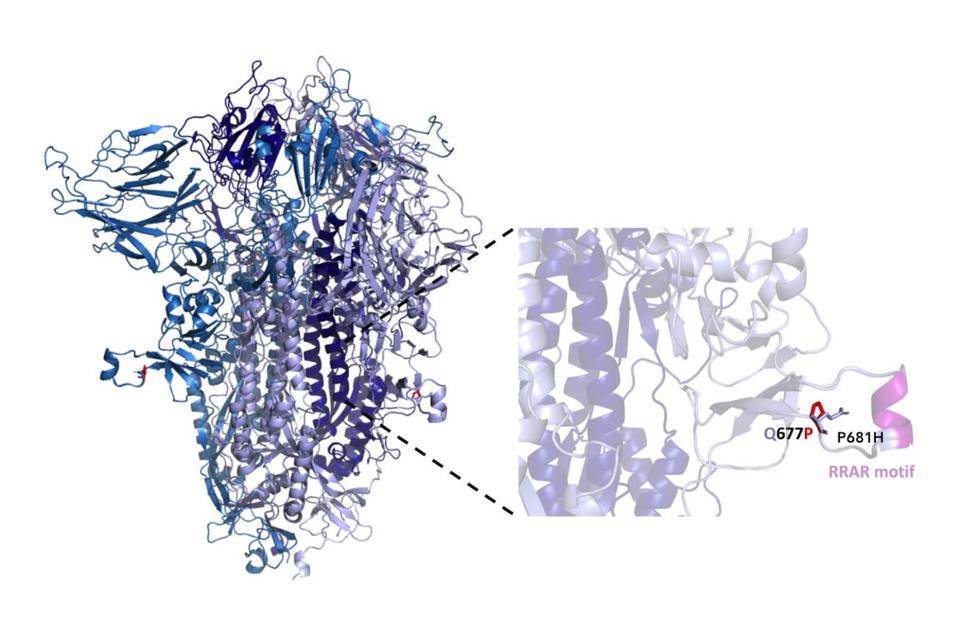
Figure 2. Structure of SARS-CoV-2 Spike protein denoting location of Q677P within a disordered loop adjacent to the furin cleavage site.
“EMERGENCE IN LATE 2020 OF MULTIPLE LINEAGES OF SARS-COV-2 SPIKE PROTEIN VARIANTS AFFECTING AMINO ACID POSITION 677” HTTPS://WWW.MEDRXIV.ORG/CONTENT/10.1101/2021.02.12.21251658V2
Though mutations at the 677 site haven’t been observed in most of the new variants spreading around Europe, South America, and elsewhere, others have that are also located in the gray area between the receptor-binding and fusion peptide domains. The B.1.1.7 (UK) variant, for instance, has a P681H mutation just a few amino acids away, while a mink variant making the rounds in Denmark has an I692V mutation in similarly close range (Figure 2). The 677 site is also not far from the furin cleavage site, located between the S1 and S2 proteins at amino acids 685 to 686. I’ve speculated in my new book Variants! that an additional furin cleavage site, which decorates the surface of the virus with spikes that are primed and ready to go, is one of the reasons SARS-CoV-2 is so infectious. It may be the case that the 677 mutations enhance this mechanism—akin to adding a hair trigger to a cocked pistol—but much more research is needed before we can say this with certainty.
In my next piece, I’ll go over why the new variants represent an opportunity for the US to reevaluate and upgrade its public health protocols. More than just preventing the worst, these measures will be a move towards the level of pandemic preparedness that will protect us from future outbreaks of Covid-19 and any other diseases to come, no matter how various they may become.
Originally published on Forbes (February 19, 2021)